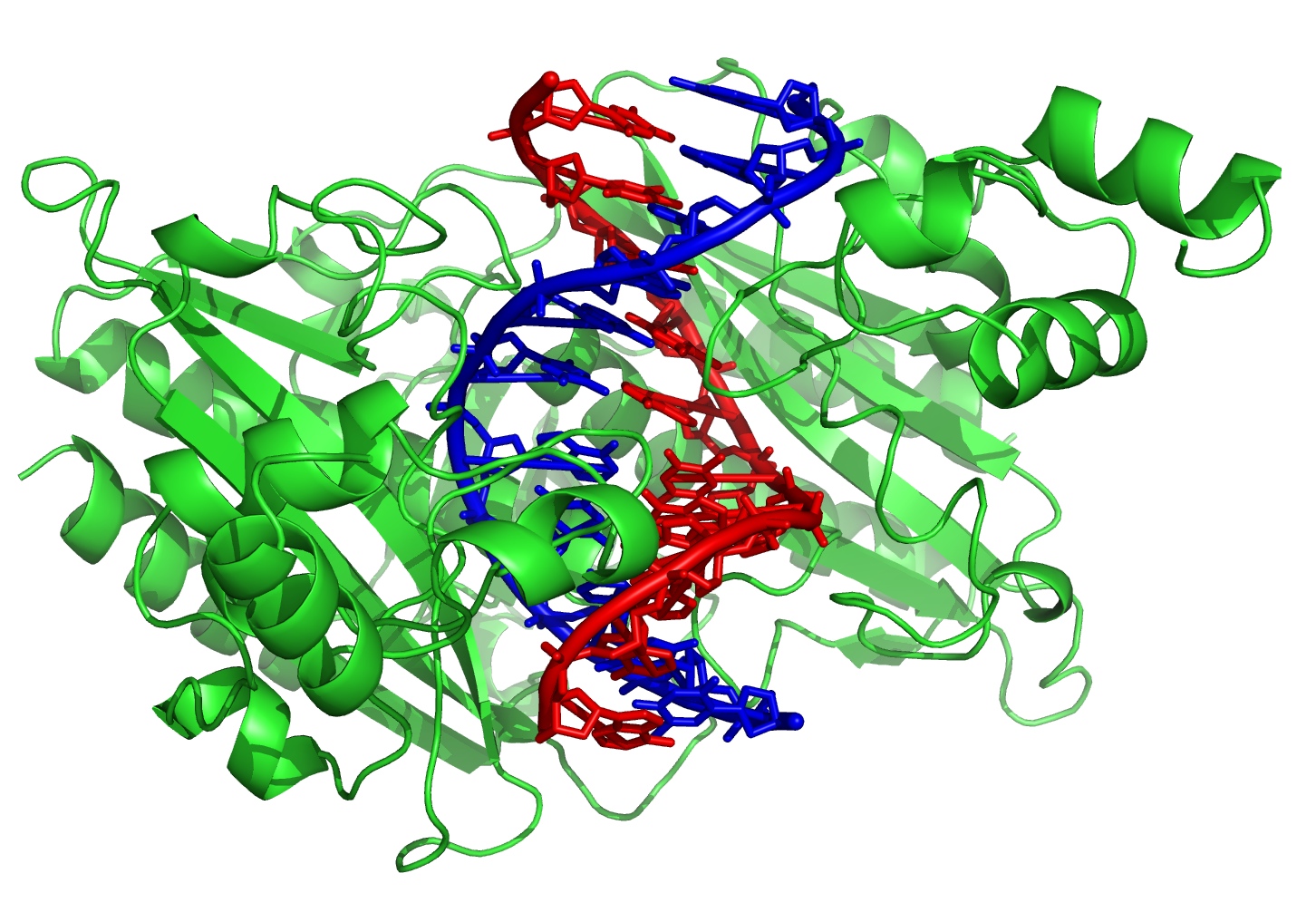
The Electric Slide
DNA may contain the parts list for building a biological cell, but it takes proteins to read it. In the 5 June Physical Review Letters, a French team reports they have found the physical mechanism by which proteins can easily slide along DNA in search of their targets. The researchers used computer simulations and analytical calculations to show that a simplified model of a protein is attracted to DNA until it gets within a half-nanometer or so. At this short range, the protein is repelled, so it can slide freely until it finds its target sequence and binds more tightly. The results provide a more complete physical picture of this critical biological process.
The proteins that interact with DNA, known as DNA-binding proteins, are essential for the operation of cells. They copy DNA, turn genes on and off, and translate genes into templates for protein production. Each protein that performs one of these functions has an affinity for a particular sequence of DNA, its target region.
To find its target, a DNA-binding protein is thought to first bind loosely to a random point on the DNA and then slide along the strand, rather than randomly binding and unbinding until it hits the right spot by luck. However, researchers have not identified the interaction that allows proteins to slide along DNA.
To study this interaction, Vincent Dahirel of the Pierre and Marie Curie University in Paris and his colleagues focused on the charges and general shapes of the proteins and DNA but ignored the details of complete atomic structures. In their Monte Carlo simulations, they modeled the DNA as a long cylinder and the protein as one of four solids–a sphere, a cylinder, or a cylinder or cube with a notch carved into one side to “mate with” the DNA. The protein diameters and lengths were all 5 nanometers; the DNA’s diameter was 2 nanometers.
The team calculated the forces as the positively-charged protein approached the negatively-charged DNA. For the cylindrical and spherical proteins, the attraction to DNA persisted as the two approached. But the researchers were surprised that the cylindrical protein with a notch was repelled when it got within a short distance, 0.1 - 0.75 nanometers of the DNA, depending on the amount of charge they assumed for the protein. The simulations showed that, as long as the protein was at the correct distance–where attraction and repulsion balanced–it was free to slide up and down the DNA strand like a puck on an air hockey table.
To explain this repulsive force, the team focused their analytical calculations on the ions dissolved in the solution that bathes biological molecules. If the DNA surface has more negative charge than the protein has positive, some positive ions from the solution are attracted to its surface. As the protein-DNA gap closes, some of these ions become trapped, creating a region of high ion concentration. Water tries to move into such regions (the effect known as osmosis), so it presses in around the edges of the interface, creating an osmotic pressure in the gap that pushes out on the protein, the team found.
To see if these proteins really do have less charge than DNA has, Dahirel and his colleagues analyzed a dataset of 77 protein structures. They found that the average DNA-binding protein has just 17% of the surface charge of DNA. For this degree of charge imbalance, their model predicts that the protein should hover half a nanometer from the DNA surface. This number agrees well with the measured structure of a DNA-binding protein from bacteria called EcoRV.
The work is “very important,” says Daan Frenkel of Cambridge University in England. “It finally clarifies how proteins can diffuse along DNA for large distances without too much friction.”
–Michelangelo D'Agostino
Michelangelo D’Agostino is a physicist and freelance science writer in Berkeley, California.