When Does a Disease Turn Epidemic?
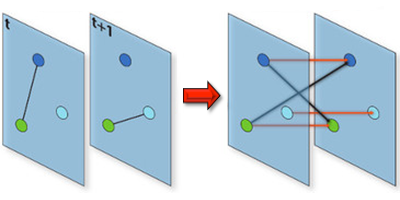
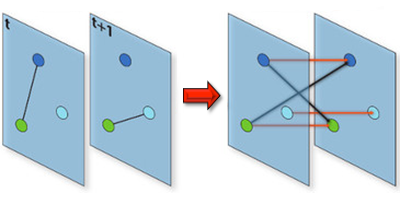
Understanding the conditions that make an infectious disease reach an “epidemic threshold” is essential for the design of efficient containment measures. Existing theories can compute this threshold only under simplifying assumptions. For instance, they often assume that the network of contacts through which the disease spreads is static. But in real-world epidemics, diseases spread between individuals in a dynamic pattern of encounters, gatherings, or travels. Vittoria Colizza of the Pierre Louis Institute of Epidemiology and Public Health of Inserm and Université Pierre et Marie Curie, France, and co-workers have now developed a model to compute analytically the epidemic threshold for networks that have arbitrary temporal dynamics.
Using tools from graph theory, the researchers expanded a well-known epidemic model so that it could describe an epidemic spreading in a network that changes in time. They showed that they could derive the epidemic threshold (the disease transmission probability above which the epidemic takes off) without assuming that the network changed in time according to a specific functional form, as existing models do. The theory allowed the researchers to accurately predict the epidemic threshold for several networks, including three time-varying networks for which empirical data were available: face-to-face encounters between researchers at a few-day conference, sexual contacts between prostitutes and their clients over the course of a year, and contact between teenagers during one day at a high-school. The formalism could describe similar “contagion” phenomena, from the dissemination of trends through social networks to the propagation of cyber viruses through the Internet.
This research is published in Physical Review X.
–Matteo Rini