Pulling on a Cell’s Strings
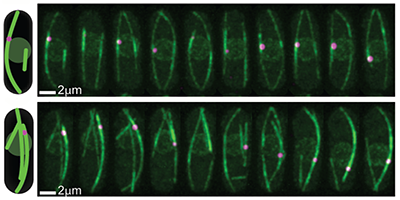
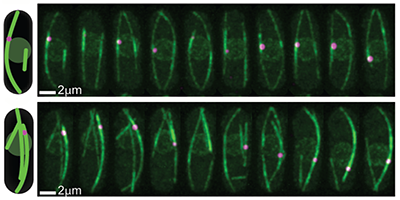
A cell’s nucleus must be centered as the cell prepares to divide to ensure the symmetric formation of two daughter cells. Tubular structures, or “microtubules,” linked to the nucleus and pushing against the cell’s walls, keep the nucleus positioned by adjusting their lengths, but it’s not clear what mechanism cues them to elongate or shorten. Now, a team led by Nenad Pavin at the University of Zagreb, Croatia, and Iva Tolić at the Max Planck Institute of Molecular Cell Biology and Genetics, Germany, has demonstrated with theory and experiments that certain motor proteins, called kinesin-8, improve the accuracy of nuclear centering by controlling the transition from a lengthening to a shortening phase of the tubules.
In the authors’ model, the nucleus’ position is determined by the pushing forces exerted by growing microtubules, which are influenced by kinesin-8 motor proteins: These walk along the microtubules and as they accumulate at the end, they can trigger a switch to a shortening phase by causing the depolymerization of the tubule’s end. The model calculates the resulting distributions of nuclear positions, predicting that the kinesin-8’s overall effect is that of balancing nuclear placement. The authors confirmed their conclusions with a series of experiments on yeast cells: First, they verified that in cells emptied of kinesin-8 the nucleus was less precisely centered (the standard deviation of its position was 70% larger). A second test displaced the nucleus with optical tweezers and found that the time for it to re-center (22 minutes) agreed with their model predictions. The new insights on the centering role of this motor protein may help researchers understand the mechanisms by which other cellular organelles are moved and positioned by microtubule forces.
–Matteo Rini
This research is published in Physical Review Letters.