How to Open a Hole in a Cell
Poking holes in a cell membrane is a standard way to deliver DNA and perform other experiments on cells. Now a team reporting in the 2 July Physical Review Letters has used computer simulations to show how the process works, at least for one hole-drilling technique. Water molecules driven into the membrane by high-frequency vibrations (ultrasound) cluster into groups that ultimately distort the membrane surface to form pores. The team identified the number of water molecules needed to generate a stable pore. Such pores may also serve as channels for drugs in future treatments.
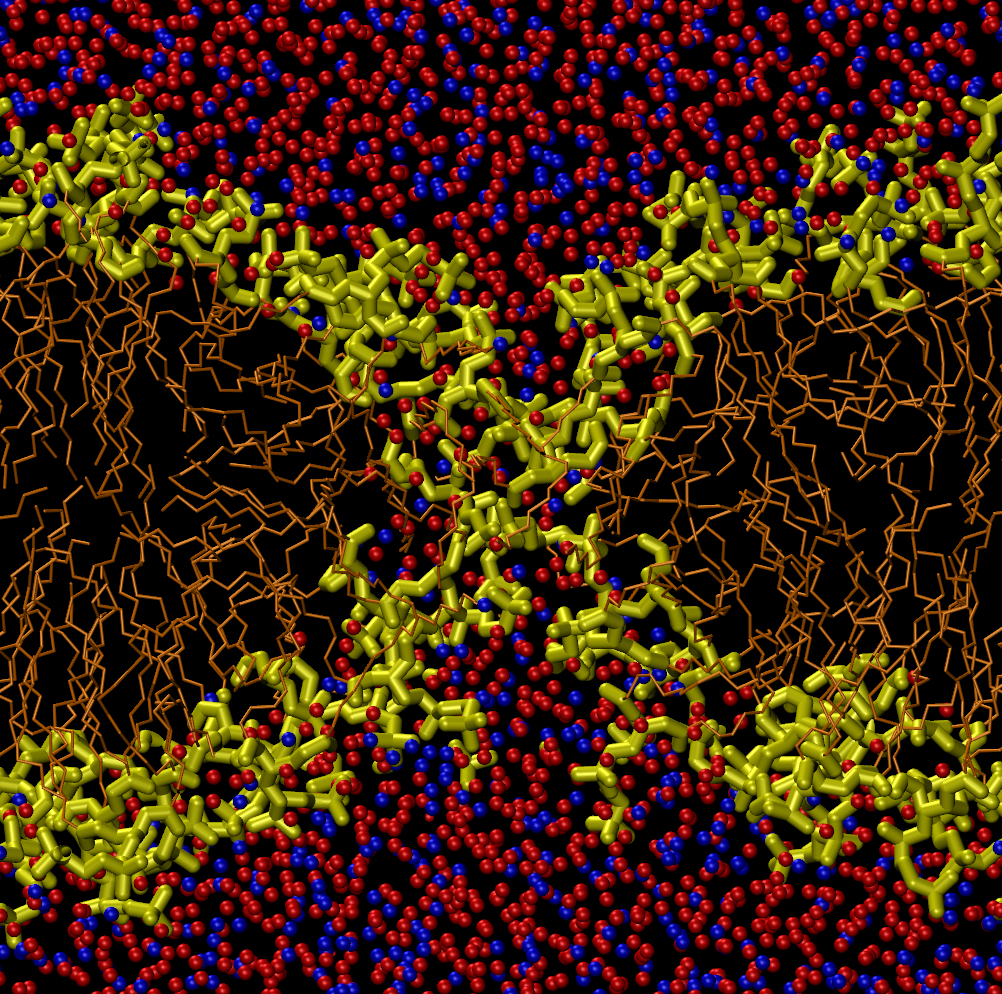
A cell membrane, the skin surrounding a biological cell, is a two-layer-thick sheet of lipid molecules. The molecules’ long, hydrophobic (water-repelling) tails are on the inside of the membrane, while their hydrophilic (water-attracting) heads face the watery solution inside and outside the cell. Electric fields, ultrasound, and other stresses can generate pores that allow water to penetrate the bilayer, a common technique in biology labs and one that may lead to new ways of delivering drugs. But the pore formation process is too fast to observe directly, so researchers have to simulate it on computers to fully understand it.
In previous simulations, Kenichiro Koshiyama of Osaka University in Japan and his colleagues studied the ultrasound “sonoporation” technique and found that it drives water molecules into the membrane, among the lipids, within picoseconds [1]. Based on these results, the team reasoned that ultrasound creates pores indirectly–if enough water molecules are present within the membrane, pores form spontaneously over the course of several nanoseconds.
To test this concept, Koshiyama and his colleagues simulated a horizontal lipid bilayer made of 128 phospholipid molecules with water molecules above and below. They started their simulation by placing between 400 and 2000 water molecules uniformly throughout the hydrophobic region of the membrane, as if driven there by ultrasound. Within several hundred picoseconds, the water molecules had clustered in the middle of the bilayer, where the density of the membrane is the lowest. This clustering was their way of minimizing contact with the hydrophobic lipid tails. The cluster then attracted the hydrophilic head groups at both surfaces of the membrane, causing surface dimples that deepened and joined to form a complete pore.
But the pore didn’t always form or remain for a long time. For clusters of 400 water molecules, the water quickly seeped out of the membrane before a pore could form, but for clusters of 800 or 1200 molecules, a pore about 1.4 nanometers in diameter usually formed. Larger water clusters created more complicated distortions in the membrane called micelles–lipid-only globs with head groups on the outside and tails on the inside. These shapes eventually formed multiple pores of less distinct shape than the single, hourglass-shaped pore created by a smaller cluster. These micelles and multiple pores were unstable and disappeared within 14 nanoseconds, whereas the hourglass-shaped pores lasted up to 100 nanoseconds, depending on the size of the initial water cluster.
When combined with the authors’ previous work, the new study gives “a plausible mechanism of sonoporation,” says membrane biophysicist Peter Tieleman of the University of Calgary in Canada. He notes that a better understanding of electrical methods for generating pores in cells has led to procedures that kill fewer cells and introduce more of the gene or drug of interest. A fuller understanding of sonoporation could be similarly useful, agrees electrical engineer Thomas Vernier of the University of Southern California in Los Angeles. “Usually it is better to understand how your method is working,” he says.
–JR Minkel
JR Minkel is a freelance science writer in New York City.
References
- K. Koshiyama, T. Kodama, T. Yano, and S. Fujikawa, “Structural Change in Lipid Bilayers and Water Penetration Induced by Shock Waves: Molecular Dynamics Simulations,” Biophys. J. 91, 2198 (2006); K. Koshiyama, T. Kodama, T. Yano, and S. Fujikawa, “Molecular Dynamics Simulation of Structural Changes of Lipid Bilayers Induced by Shock Waves: Effects of Incident Angles,” Biochim. Biophys. Acta 1778, 1423 (2008)