Bacteria Evolve to Go Against the Grain
Since Alexander Fleming discovered in 1928 that a substance secreted by a mold could kill bacteria, we have become used to the ease of administering cocktails of antibiotics to fight bacterial infections. However, the use and misuse of antibiotics in human medicine and livestock farming over many decades has had the serious side effect of selecting for bacteria that survive drug attack: many bacterial strains have emerged that are resistant to antibiotics, most famously the multi-drug-resistant Staphylococcus aureus strain, the source of Staph infections, that represents a major threat to hospital patients. The pool of effective antibiotics is running out, and finding new strategies to prevent the rapid evolution of drug resistance has become a pressing challenge for global health.
Now, two independent theoretical studies, one published in Physical Review Letters by Philip Greulich at the University of Edinburgh, UK, and colleagues [1], and the other by Rutger Hermsen at the Center for Theoretical Biological Physics, San Diego, and colleagues in Proceedings of the National Academy of Sciences [2], address how the evolution of drug resistance can be profoundly influenced by how drugs are distributed in biological tissues. Once administered, drugs do not spread evenly throughout the human body, which is compartmentalized and composed of tissues that have different affinity for retaining the drugs. The new research suggests that variations in the drug concentration may play an important role in selecting bacterial strains that are able to survive exposure to antibiotics.
The rapid evolution of drug resistance is an impressive demonstration of the efficiency of bacterial adaptation. Bacterial populations are large and cells are able to replicate rapidly (sometimes in minutes) and to exchange genetic material. These are ideal conditions for Darwinian evolution to act: slight changes, or mutations, in the genomes arise by chance from one bacterial generation to the next. While most of these changes are deleterious, some, such as resistance to an antibiotic substance, may confer an advantage to bacteria living in human tissues, allowing the population of resistant mutant strains to rapidly increase. As a consequence, unless a bacterial infection is completely wiped out by a large dose of antibiotics, drug resistant strains emerge.
Yet when a mix of antibiotics is administered, the buildup of drug resistance requires multiple mutations and becomes harder to explain [3]; in this case, the bacterial population should be driven to extinction, unless a resistant multimutant arises. This, however, becomes prohibitively unlikely for just a handful of required mutations. Why then does drug resistance evolve so readily?
The work of Greulich et al. and Hermsen et al. could solve this puzzle because it demonstrates that spatial variations in drug concentrations allow for a gradual selection of drug resistant mutants. Heterogeneities in drug concentrations have been previously recognized as a potential driver of drug resistance [4,5], as was demonstrated in a recent microfluidic experiment [6]. Yet quantitative models have been lacking so far. The new theoretical studies fill this gap and help identify parameters that may be key to controlling drug resistance evolution.
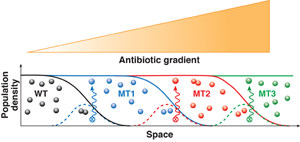
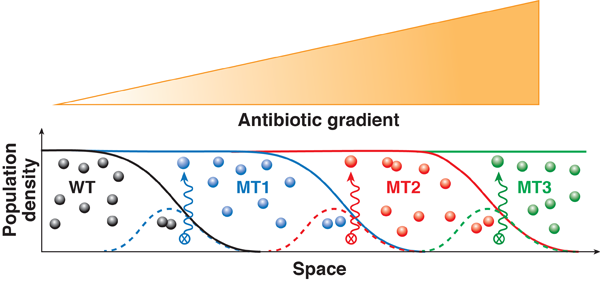
In a gradient of drug concentration, resistance can evolve following the stepwise dynamics illustrated in Fig. 1. Initially, only a small population of wild-type (i.e., nonmutant) cells can be sustained in the regions where drug concentration is low. The growth of bacteria in this suitable habitat is balanced by the death of bacteria that move by chance into less hospitable regions of higher drug concentration. During this continual population turnover, a mutant will eventually appear that is slightly more resistant than the wild type. This mutant will then be able to endure higher drug concentration and move up the concentration gradient. Thereby the mutant population can overgrow the wild type and reach a steady-state distribution that covers a slightly larger region than the original wild type. Then the process stalls until the population turnover generates a new mutant with an additional beneficial genetic change. This next-generation mutant will colonize a larger area of higher drug concentration and eventually supersede the previously fittest mutant. By this mechanism, the bacterial population can plausibly penetrate the concentration gradient and efficiently develop any number of mutations provided that each mutation increases resistance.
The two research groups consider how this Darwinian selection process can work in two different regimes. Greulich et al. assume a relatively smooth gradient in drug concentration, and analyze the resulting dynamics in a spatially continuous one-dimensional framework [1]. Their analysis leads to predictions for the drug resistance buildup time depending on few control parameters, such as bacterial densities, the steepness of the concentration gradient, mutation rates, and diffusion coefficients. The authors demonstrate that heterogeneities indeed enable a specific mechanism of rapid evolution of drug resistance. They also show that drug gradients speed up drug resistance evolution only if resistance strictly increases in each step of the mutational path leading from the original wild type to the drug resistant mutant. In contrast, if an intermediate mutant is less resistant, gradients may in fact slow down the evolution of resistance. Rutger Hermsen and colleagues have developed a different model, based on an analogy to ecological models characterized by habitat heterogeneity [7], in which the bacterial population is fragmented in relatively isolated subpopulations.
The two analyses hinge on the question of how new resistant mutations arise and take over the leading edge of the current bacterial population. Hermsen and colleagues assume that the heterogeneous drug distribution leads to sharp spatial gradients in bacterial growth rates. In this case, new mutations typically appear in densely populated areas where they do not enjoy a direct selective advantage. To grow and take over the whole population, such mutants must also migrate into the region of higher drug concentration where they have a growth rate advantage compared to the currently dominating strain. This “mutate and migrate” mechanism is in contrast to the work by Greulich and colleagues, who focus on the case of relatively shallow gradients. There, mutations that take over usually appear in regions where they already enjoy a direct growth rate benefit, and there is no need to wander into a different region in order to establish themselves, although migration subsequently allows these successful mutants to expand the population’s range.
Which mechanism is the dominating one in a given biological scenario? If mutants emerge where they enjoy a direct benefit, mutations will grow where they arise, as assumed by Greulich and colleagues. By contrast, if antibiotic gradients are sharp or if bacteria show a mean drift direction towards the antibiotics, possibly due to blood flow or chemotaxis (i.e., movement driven by the concentration of certain chemicals), the mutate and migrate mechanism of Hermsen and collegues may control the buildup of antibiotic resistance. Recent advances in the theory of wavelike expansion of populations may help develop a unified model that captures both mechanisms [8,9].
Experiments in microfluidic devices along the lines of Ref. [6] or in petri dishes [10] may now be devised to test the basic trends predicted by the two new studies. Further theoretical work is needed to take into account more than one spatial dimension, realistic heterogeneities in drug concentrations, or the specific mutational pathway leading to resistance. Yet the simplicity of these two models, based on only few assumptions, makes them quite general and an excellent starting point for quantifying and understanding the development of drug resistance in a broad set of systems, including populations of cancerous cells developing resistance against chemotherapeutic agents.
References
- P. Greulich, B. Waclaw, and R. J. Allen, ”Mutational Pathway Determines Whether Drug Gradients Accelerate Evolution of Drug-Resistant Cells,” Phys. Rev. Lett. 109, 088101 (2012)
- R. Hermsen, J. B. Deris, and T. Hwa, “On the Rapidity of Antibiotic Resistance Evolution Facilitated by a Concentration Gradient,” Proc. Natl. Acad. Sci. 109, 10775 (2012)
- M. Lipsitch, “The Rise and Fall of Antimicrobial Resistance,” Curr. Trends Microbiol. 9, 438 (2001)
- T. B. Kepler and A. S. Perelson, “Drug Concentration Heterogeneity Facilitates the Evolution of Drug Resistance,” Proc. Natl. Acad. Sci. 95, 11514 (1998)
- F. Baquero, M. C. Negri, M. I. Morosini, and J. Blázquez, “Antibiotic-Selective Environments,” Clin. Infect. Dis. 27, Suppl. 1, S5 (1998)
- Q. Zhang, G. Lambert, D. Liao, H. Kim, K. Robin, C. Tung, N. Pourmand, and R. H. Austin, “Acceleration of Emergence of Bacterial Antibiotic Resistance in Connected Microenvironmentsm,” Science 333, 1764 (2011)
- R. Hermsen and T. Hwa, “Sources and Sinks: A Stochastic Model of Evolution in Heterogeneous Environments,” Phys. Rev. Lett. 105, 248104 (2010)
- R. Lehe, O. Hallatschek, and L. Peliti, “The Rate of Beneficial Mutations Surfing on the Wave of a Range Expansion,” PLoS Computational Biology 8, 1002447 (2012)
- R. A. Neher and B. I. Shraiman, “Statistical Genetics and Evolution of Quantitative Traits,” Rev. Mod. Phys. 83, 1283 (2011)
- K. S. Korolev, M. J. I. Müller, N. Karahan, A. W. Murray, O. Hallatschek, and D. R. Nelson, “Selective Sweeps in Growing Microbial Colonies,” Phys. Biol. 9, 026008 (2012)