Coherent Signal Picks Out Biomolecular Interactions
Proteins rarely act alone: Their functioning requires that they establish contact with other proteins and molecules, mostly through noncovalent interactions (i.e., interactions that do not involve the sharing of electrons, such as hydrogen-bond and van der Waals interactions). The study of such interactions is key for the understanding of biology at the molecular level, and may have important implications for drug discovery or the development of diagnostic tests. It is thus crucial to develop measurement techniques that can selectively probe these interactions, characterizing, for instance, the formation rate and strength of a specific protein-ligand complex, while discriminating from other, nonspecific interactions (like those arising, for instance, between a protein and fluctuating solute molecules). Writing in Physical Review X, Christof Fattinger, of Roche Innovation Center, Basel, Switzerland, investigates theoretically a novel analytical method, called focal “molography” (molecular holography [1]), which represents a potential breakthrough for the selective detection of molecular interactions [2].
Noncovalent interactions play a crucial role in biology. They determine the D structure of biopolymers and are important in vital processes like signal transduction, metabolism, DNA replication, the binding of drugs to receptors, and immune responses. The ability to characterize these interactions and observe their evolution in real time is thus essential for the study of the kinetics of such processes. Compared to the covalent bindings that hold molecules together, noncovalent forces are much weaker, comparable with thermal fluctuations. This is what makes them so ubiquitous in biology: They enable the formation of a dynamic network of interacting molecules, kept together by bonds that can form and break again. But what makes them important also poses the main challenge to their study: it’s hard to find spectroscopic observables (like an electronic or vibrational transition) with which to characterize them and they are susceptible to fluctuations of the environment. Furthermore, many other interactions and nonspecific interactions occurring on similar energy scales may complicate the experiments.
Only a few methods exist that can probe noncovalent interactions in material samples of less than a microgram —a key requirement for noninvasive diagnostic tests or the study of expensive, newly discovered proteins. Examples include UV-visible spectrophotometry [3], atomic force microscopy (AFM) [4], electrospray ionization mass spectrometry (ESI-MS) [5], matrix-assisted laser desorption/ ionization mass spectrometry (MALDI-MS) [6], and biomolecular interaction analysis involving optical biosensors [7,8]. Most of these methods rely on the statistical analysis of large ensembles of molecules, and are thus often limited by background noise produced by nonspecific bindings. In other cases (e.g., AFM), the observation focuses on individual molecules: this makes nonspecific bindings easier to exclude from the data collection. But even in AFM, the environment of the investigated molecules is usually not completely under control, representing a potential source for noise. Further, just measuring a single event—in particular for larger molecules—is usually not enough as in most cases it is not exactly known what has been measured. Therefore, single-molecule investigations require both repetitive measurements and statistical analysis. But performing a series of highly sensitive single-molecule measurements on a very large number of molecules remains challenging. Hence, an important goal for biomolecular-interaction detection is to avoid major sources of noise due to nonspecific bindings, while enabling the collection of data sets of sufficient sizes to be statistically relevant. The new molography technique proposed by Fattinger addresses this task in a new and powerful way.
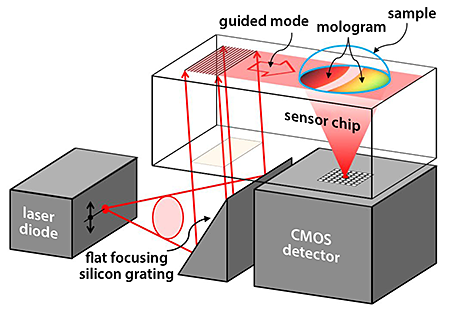
Fattinger’s design (see Fig. 1) combines state of the art lithography, molecular self-assembly and optical technology into one powerful technique. The main idea behind focal molography is to construct a clever geometrical arrangement in which molecular binding sites on a monolithic substrate are spatially ordered on scales that match the wavelengths of coherent light, allowing probing of the molecule-reactant pairs with ultrasensitive diffractive methodologies. In the scheme, one interaction partner (the “receptor,” e.g., a specific protein) of the probed biospecific interaction is linked to a micropatterned area engineered on the top surface of a thin-film optical waveguide. The linked receptors form the so-called “mologram”: a coherent submicron assembly of receptor molecules with active binding sites. The receptor molecules in the mologram are immobilized on curved lines with intermolecular distances smaller than the light wavelength, but much larger than a single receptor molecule. This serves a twofold purpose. First, it allows sensitive coherent probing of the many molecules contained in a mologram. Second, it prevents so-called “steric hindrance” effects in which a conformational change of a molecule is hindered by its proximity to other molecules.
Fattinger imagines that a thin film of high-refractive-index material ( ) could be used as an optical waveguide to strongly couple laser light to the molecular film. (A reflective-imaging silicon grating [9] would be used to convert the diverging laser beam from a laser diode into a parallel beam.) The evanescent field of the guided mode extends into the thin (few nm to nm) mologram, probing the molecules. While propagating through the mologram, the guided mode partially decouples from the waveguide through diffraction by the molecules in the mologram. The coherently assembled molecules focus the diffracted light into a diffraction-limited “pencil beam” that is collected by a detector. The coherent light intensity in the focus of the mologram can be related to refractive-index changes in the nanoenvironment of the probed molecules in the mologram, from which information on the molecular interaction is deduced: The index-of-refraction change is related to the number of bindings and can thus be used to measure the probability for such a binding to take place under given conditions.
The scheme provides signal amplification thanks to the fact that the phase contrast is enhanced by the coherent addition of signals from orderly assembled molecules. This brings two unique advantages over existing optical sensing methods. (i) It eliminates the effect of nonspecific binding, since nonspecific bindings are not arranged in the same spatially ordered way as the receptors, and the light they diffract does not contribute to the detected signal. (ii) It has a very high sensitivity that can be improved (virtually without limits) by increasing the size of the sensor. Since the sensitivity of focal molography arises from coherent interference effects, it scales with the square of the diameter of the mologram. The diameter of the mologram proposed by Fattinger lies in the -micrometer to -millimeter range. Larger molograms would allow better sensitivity but also require higher precision in the lithographic fabrication process to enable diffraction-limited focusing.
The readout of a mologram on a chip by a small optoelectronic system could be integrated into a small analytical device, as illustrated in Fig. 1. Such a compact device may be suitable for self-testing or point-of-care-testing of analytical or diagnostic parameters.
Focal molography requires only a minuscule amount of bound biological matter for a measurement (Fattinger envisions femtograms of coherently assembled molecules at the mologram’s binding sites [2]). Other advantages include the possibility of probing biomolecular interactions in aqueous solution (the natural environment in which they occur); the ability to deliver results in a short time (beneficial for diagnostic tests); and the ability to observe the progression of a molecular interaction in real time—essential for the measurement of the kinetics of a biomolecular interaction.
The new method will not make single-molecule measurements obsolete; rather, it will complement them. One could, for example, envision combining AFM and focal molography in one microscopic system that measures multiple parameters of biomolecular interactions on the same chip: the molographic readout could detect the onset and the progression of a molecular interaction, while AFM could detect structural details of the molecular-interactions that can’t be resolved by optical means.
Focal molography might find important applications in the design of immunoassays— biochemical analyses that are the basis of many clinical tests, including prostate cancers, heart disease, and sport antidoping tests. Immunoassays measure the concentration of a given macromolecule (the “analyte”) by revealing its binding with a receptor (e.g., an antibody) through a detectable “label” (e.g., a gold or latex nano particle [10]). At the typical low concentration of the analyte in the sample, only about in receptor molecules becomes occupied by an analyte molecule [10,11]. Under these conditions, nonbiospecific binding and the size of the sensor are the key limiting factors for detection [11,12]. Focal molography schemes, optimized for given analyte-receptor combinations, might thus boost the sensitivity of a number of life-saving diagnostic methods.
References
- The term “molography” was used for the first time in this context in a discussion between Christof Fattinger and Klaus-Peter Stengele. Fattinger suggested calling his analytical method “molography,” combining “molecule” and “holography” in one word. A mologram is a synthetic hologram that is composed of molecules with interaction sites. Focal molography probes a molecular interaction in the mologram by monitoring diffracted light in a diffraction-limited focus. (The discussion between K.P. and C.F. took place on a bench in the Platzspitz Park behind the Swiss National Museum in Zurich, Switzerland on August 9, 2012.)
- Christof Fattinger, “Focal Molography: Coherent Microscopic Detection of Biomolecular Interaction,” Phys. Rev. X 4, 031024 (2014)
- K. Hirose, “A Practical Guide for the Determination of Binding Constants,” J. Inclusion Phenom. Macrocyclic Chem. 39, 193 (2001)
- Dynamic Force Spectroscopy and Biomolecular Recognition, edited by A. R. Bizzarri and S. Cannistraro (CCR Press, Boca Raton, USA, 2012)
- B.Ganem, Y.-T. Li, and J.D. Henion, ”Detection of Noncovalent Receptor–Ligand Complexes by Mass Spectrometry,” J. Am. Chem. Soc. 113, 6294 (1991)
- M. Karas, U. Bahr, A. Ingendoh, and F. Hillenkamp, “Laser Desorption/Ionization Mass Spectrometry of Proteins of Mass 100 000 to 250 000 Dalton,” Angew. Chem., Int. Ed. Engl. 28,760 (1989)
- R. Karlsson, A. Michaelsson, and L. Mattsson, “Kinetic Analysis of Monoclonal Antibody-Antigen Interactions with a New Biosensor Based Analytical System,” J. Immunol. Methods 145, 229 (1991)
- M. A. Cooper, “Optical Biosensors in Drug Discovery,” Nat. Rev. Drug Discovery 1, 515 (2002)
- C. M. Greiner, D. Iazikov, and T. W. Mossberg, “Diffraction-Limited Performance of Flat-Substrate Reflective Imaging Gratings Patterned by DUV Photolithography,” Opt. Express 14, 11952 (2006)
- S. Kubitschko, J. Spinke, T. Bruckner, S. Pohl, and N. Oranth, “Sensitivity Enhancement of Optical Immunosensors with Nanoparticles,” Anal. Biochem. 253, 112 (1997)
- R. P. Ekins, “Ligand Assays: From Electrophoresis to Miniaturized Microarrays,” Clin. Chem. 44, 2015 (1998)
- T. M. Squires, R. J. Messinger, and S. R. Manalis, “Making It Stick: Convection, Reaction and Diffusion in Surface-Based Biosensors,” Nature Biotechnol. 26, 417 (2008)