Measuring Membrane Mechanics
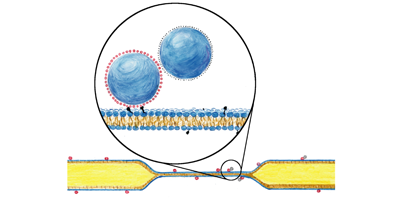
Lipid bilayers—flexible two-dimensional fluids of proteins and lipids—govern the dynamics of cell membranes. Understanding the dynamics of these bilayers is important if we want to explain their biological functions. For example, the membrane viscosity sets the time scales for protein diffusion, assembly of signaling complexes, and other processes that rely on two-dimensional motion. Measuring membrane viscosity, however, is notoriously difficult. Writing in Physical Review Letters, Raghuveer Parthasarathy and colleagues from the University of Oregon, Eugene, describe a new technique that can be used to accurately obtain membrane viscosity by probing the diffusion of anisotropic lipid-anchored particles.
Tracers made of paired 100-nanometer-diameter spheres were bound to particular lipids, which were then incorporated into the membrane. The location and orientation of the tracers were found from fluorescence images allowing measurement of both translational and rotational Brownian motion, the combination of which allows the two-dimensional viscosity to be determined. Since the technique only requires the binding of tracers to lipids, it can be applied to a wide variety of membrane systems from simple bilayers to more complex multilayer systems and vesicles.
Using this approach, the authors measured the change in membrane viscosity induced by a particular cargo trafficking protein that, in cells, helps to shape the membranes into curved buds. Surprisingly, they found that this particular protein increased membrane viscosity by more than an order of magnitude. Their results open the door to studying a wide array of mechanical effects induced by individual membrane-active proteins. – Katherine Wright