It’s All in the Sequence
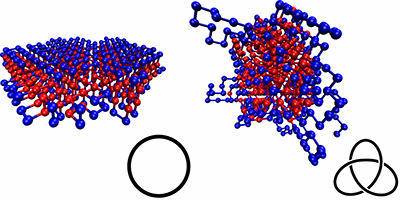
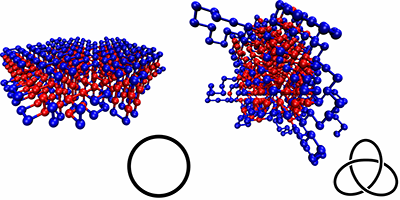
Everyday experience shows that strings easily knot. Preventing this requires careful folding or winding when stowing away. Molecular ropes, like polymer chains, can suffer the same fate, but that is not true for biopolymers like proteins and DNA; despite their complex folded conformations, they rarely get knotted. A new study by Thomas Wüst from the Swiss Federal Institute of Technology (ETH), Zurich, and colleagues suggests the differences in the interactions between different parts of the chain of a protein (due to the sequence of amino acids forming it) is what controls and prevents knotting.
Wüst et al. simulated simplified, coarse-grained proteins made of 500 monomers (“residues”), which were either hydrophobic or polar. The authors compared different types of sequences: homoresidue chains, randomly ordered ones, and chains designed with specific repetition patterns, calculating their various ground-state conformations and checking for knots. They found that the knottiness of the chain depended on the sequence, and they were able to design sequences that were either highly knotted or almost completely knot-free. Sequences that were free of knots typically produced neatly folded, locally ordered structures, with none of the extended loops seen in the knotted sequences.
The proteins and sequences investigated here are much simpler than real proteins, which are made of twenty amino acids, rather than two. However, the authors speculate that sequence could have been a controlling factor in the evolution of proteins, allowing them to evolve towards knot-free conformations that can reliably perform their functions.
This research is published in Physical Review Letters.
–Katherine Wright