Microbial Expansion Shaped by Fluid Flows
Life is shaped by physics. Flow physics to be specific. It is now uncontested among scientists that early forms of life originated in water around four billion years ago and developed into single-celled microorganisms from which all life evolved. The fluid environment plays a central role in the morphology of these microorganisms both at individual and collective levels. New insight into the effects of flow physics on microbial colonies comes from a study by Severine Atis, from Harvard University, and her colleagues [1]. The authors combined carefully designed laboratory experiments with elegant mathematical modeling to show that the metabolic activity of nonmotile cells growing on liquid surfaces induces large-scale flows. These flows, in turn, direct the territorial expansion and genetic properties of the colony. Such metabolically induced flows could have a role in the transport of microorganisms in the human body.
Many types of organisms grow on liquid surfaces. A familiar example is pond scum, which is made up of algae that thrive in stagnant water conditions. However, growing cells on liquid surfaces in controlled laboratory experiments is no easy feat. Most studies examine growth on hard (agar) substrates [2] or in bulk fluids [3]. Nonmotile cells growing on an agar surface rely on molecular diffusion to feed. In this paradigm, cells consume nutrients locally, creating a depletion of nutrients in their vicinity and inducing, by Fick’s law of diffusion, a flux of nutrients from the surroundings towards the colony [4]. The rate of nutrient in-flow scales linearly with the colony’s radius, whereas the rate of nutrient consumption scales with the area of the colony, or the radius squared. Therefore, once a colony reaches a certain size, diffusion becomes insufficient to sustain cells in the interior region, and only cells on the edge have enough nutrients to grow and divide. This edge-dominated growth produces a linear expansion rate of the colony’s radius. In addition, different strains of the microorganism will be localized in separate sectors along the edge, a phenomenon often referred to as “genetic demixing” [5].
Atis and colleagues explored what happens on a liquid substrate in which nutrients can be transported via fluid flows. For their liquid substrate, they chose a nutrient-laden fluid whose viscosity was controlled by introducing a cellulose polymer. The viscosity level was made high— 104 to 105 times that of water—with the aim of confining microbial growth to the liquid-air interface. Such high-viscosity liquid is similar to the nasal, lung, and cervical mucus secretions of humans, which act as a trap to invading microorganisms.
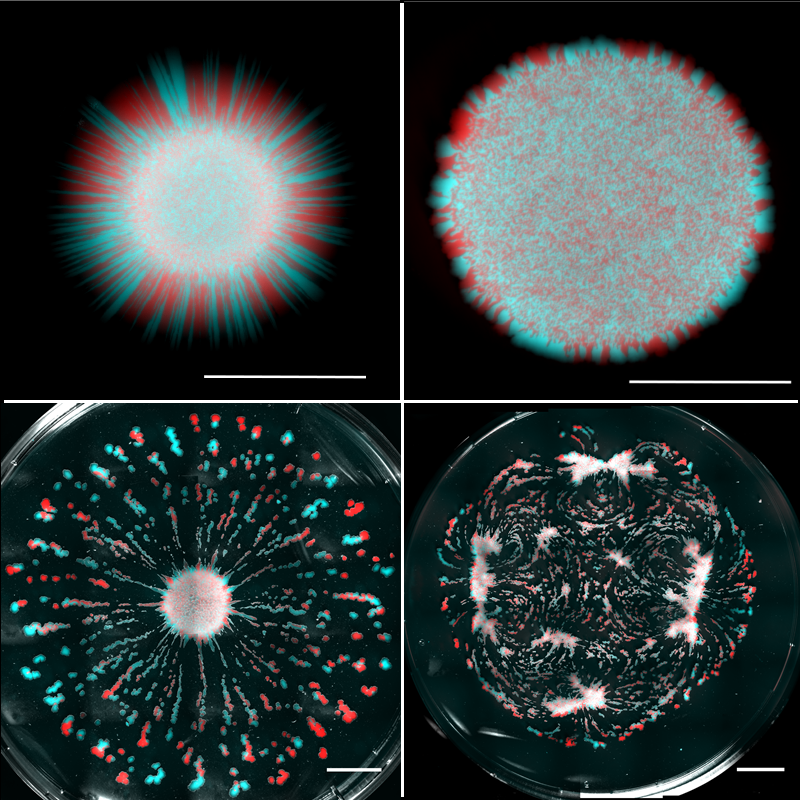
Atis et al. used their liquid substrates to grow initially well-mixed populations of two strains of nonmotile yeast cells (Saccharomyces cerevisiae). The underlying fluid was initially static—any flows that developed were a result of internal dynamics. The strains were each labeled with a different fluorescent marker—one colored red in images and the other blue—which allowed their expansions to be spatially discriminated. Atis et al. systematically lowered the liquid viscosity and compared the colonies’ spatial organization to those grown on nutrient-rich agar (Fig. 1). On agar, as previously observed, only cells dividing near the edge contributed to the colony expansion, leading to linear growth and genetic demixing—visible as separate red and blue regions along the outer edge. On high-viscosity liquid substrates, Atis et al. observed circular colonies similar to those grown on agar, but they were larger because of an initial stage of exponential growth caused by cell division throughout the entire colony. At intermediate viscosities, the colonies developed monoclonal “fingers” of a single strain type. The behavior changed dramatically at the lowest viscosities, where the expanding colonies broke up into many mutually repelling fragments that were “genetically diverse” in the strains they contained.
The results showed that different modes of colony expansion exist, depending on the liquid properties of the growth substrate. To understand how viscosity in the underlying fluid affects colony behavior on the surface, Atis and colleagues tracked fluid motion using florescent beads mixed in with the liquid. What they found is indeed fascinating: the metabolic activity of the colony induces, through a gradient in the nutrient density, fluid flows in the liquid substrate at speeds that far exceed the expansion rate of the colony. These flows, in turn, alter both the colony morphology and the spatial distribution of the different yeast strains.
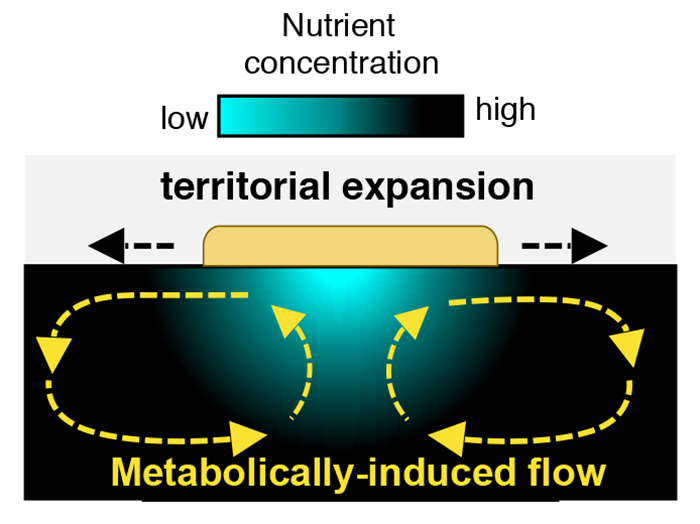
The presence of substrate flows is a clear break with the diffusion-based paradigm that defines microbial expansion on agar surfaces. Using simulations, Atis and colleagues modeled the mechanisms that produce these flows. Nutrient depletion reduces the density of the fluid medium itself, causing a density gradient in the bulk fluid below the colony. In the confined geometry of the petri dish, that density gradient combined with depth-related pressure differences leads to two large-scale, counterrotating vortices with fluid velocities up to 10 times larger than the radial expansion rate of the colony (Fig. 2).
These flows have two important functions. First, they replenish the nutrient-depleted zone by transporting nutrient-rich liquid to the colony, allowing cells in the interior region of the colony to grow and reproduce. Second, these flows create outward radial shear forces along the colony’s surface that can transport cells in the radial direction. Indeed, because the viscosity of the liquid impacts the intensity of the metabolically induced flows, the territorial expansion of the colony can result in dispersal of single-strain clusters for intermediate viscosities or genetically diverse islands for low viscosities (Fig. 1). However, the viscosity is just one control parameter: this type of expansion mode will also depend on the colony’s metabolic rate. We can therefore imagine a situation where two microbial species on the same substrate may expand in distinct ways because of differences in their metabolic rates.
The flows discovered by Atis et al. help deliver nonmotile microorganisms to new territories, which would have been inaccessible on a hard or highly viscous substrate. These new modes of dispersion could have significant implications for understanding the survival, maintenance, and evolution of microbial genotypes in various aquatic and ecological environments, including in the human body. For example, one could envision the use of this new culture-based system to study populations of beneficial and pathogenic microbes that live at air-mucus interfaces in respiratory airways [6]. Researchers may also want to revisit earlier bacterial experiments that were performed on agar. For example, bacteria engage in intercellular communication and competition through diffusible molecules [7] or direct cell-cell contact [8, 9], which can result in genetic demixing [10]. Flow fields generated on liquid surfaces could transport these molecules and cells to alter interaction outcomes. Therefore, as Atis et al. have shown, metabolically induced flows may contribute to such interactions in a way that was missed.
This research is published in Physical Review X.
References
- S. Atis, B. T. Weinstein, A. W. Murray, and D. R. Nelson, “Microbial range expansions on liquid substrates,” Phys. Rev. X 9, 021058 (2019).
- H. Berg, Random Walks in Biology (Princeton University Press, Princeton, 1993)[Amazon][WorldCat].
- O. Hallatschek, P. Hersen, S. Ramanathan, and D. R. Nelson, “Genetic drift at expanding frontiers promotes gene segregation,” Proc. Natl. Acad. Sci. U.S.A. 104, 19926 (2007).
- R. Rusconi, J. S. Guasto, and R. Stocker, “Bacterial transport suppressed by fluid shear,” Nat. Phys. 10, 212 (2014).
- M. Matsushita, F. Hiramatsu, N. Kobayashi, T. Ozawa, Y. Yamazaki, and T. Matsuyama, “Colony formation in bacteria: Experiments and modeling,” Biofilms 1, 305 (2004).
- J. Nawroth, E. Kanso, A. Ryan, and A. van der Does, (to be published).
- M. Whiteley, S. P. Diggle, and E. P. Greenberg, “Progress in and promise of bacterial quorum sensing research,” Nature 551, 313 (2017).
- R. M. Stubbendieck and P. D. Straight, “Multifaceted interfaces of bacterial competition,” J. Bacteriol. 198, 2145 (2016).
- V. Troselj, A. Treuner-Lange, L. Søgaard-Andersen, and D. Wall, “Physiological heterogeneity triggers sibling conflict mediated by the type VI secretion system in an aggregative multicellular bacterium,” mBio 9, e01645 (2018).
- L. Speare, A. G. Cecere, K. R. Guckes, S. Smith, M. S. Wollenberg, M. J. Mandel, T. Miyashiro, and A. N. Septer, “Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host,” Proc. Natl. Acad. Sci. U.S.A. 115, E8528 (2018).