How a Virus Rolls Itself Across a Cell Surface
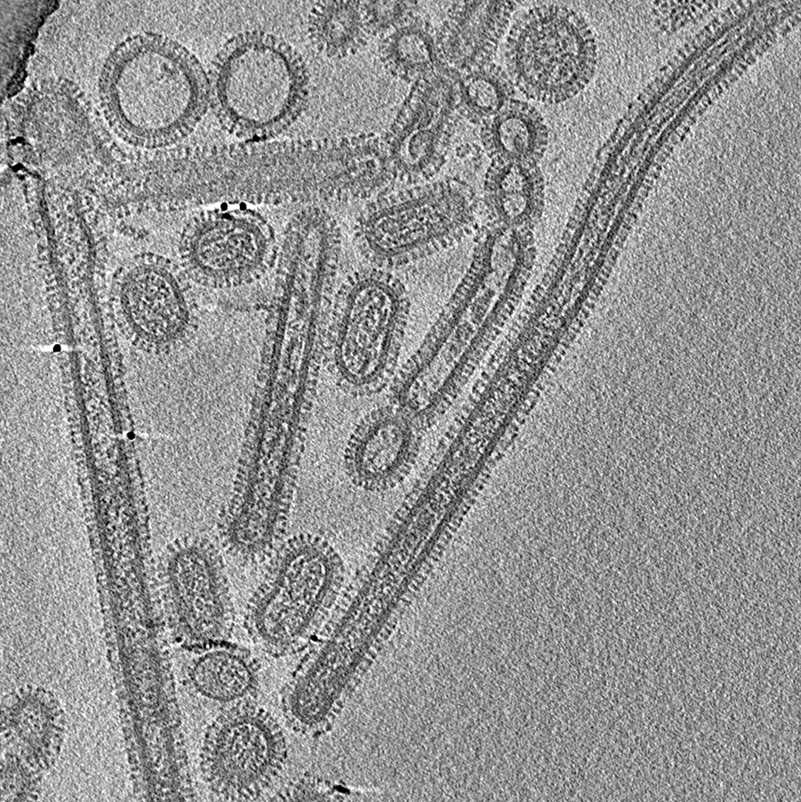
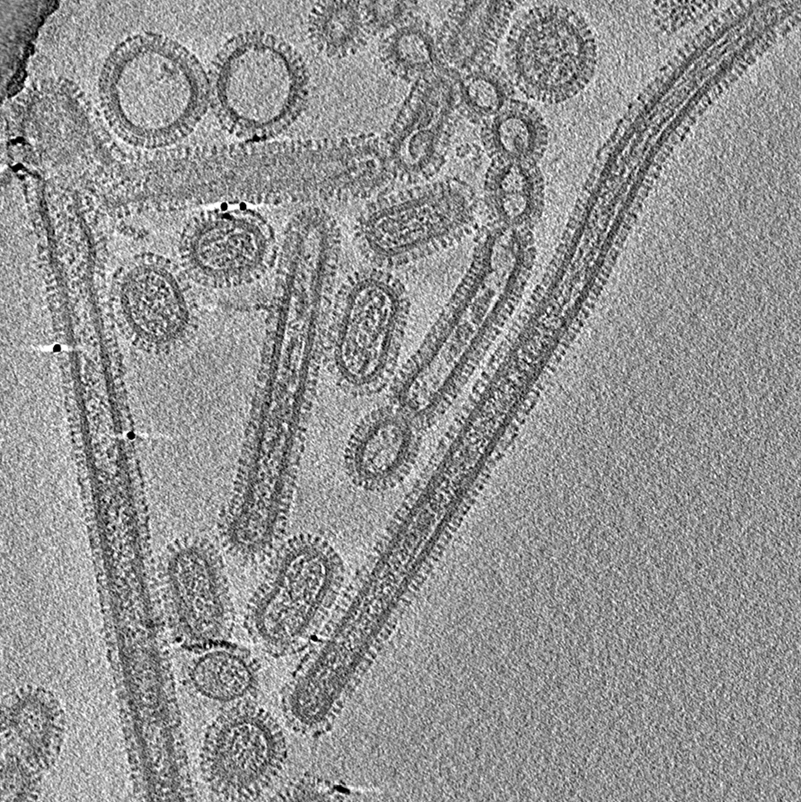
Before an influenza virus invades a cell in the airway, it stretches into a filamentary structure that rolls across the cell’s surface like a pencil on a desktop. Two theorists now propose a new mechanism for the rolling [1]. They show that two proteins on the viral surface act together to propel the motion through interactions with the cell membrane. Understanding this process might point to ways of preventing infection. However, some experts will not be convinced without more detailed modeling.
Why the influenza virus rolls isn’t known. One possibility is that this motion achieves a good compromise between allowing the virus to seek a point of attack and keeping it firmly attached to the membrane via chemical links—rather like the crampon spikes that a mountaineer uses to stay anchored to ice while crossing it. The lung tissues that the virus targets are lined with tiny appendages called cilia that move vigorously to sweep away dirt and pathogens, so the virus needs a good hold to avoid getting flushed away.
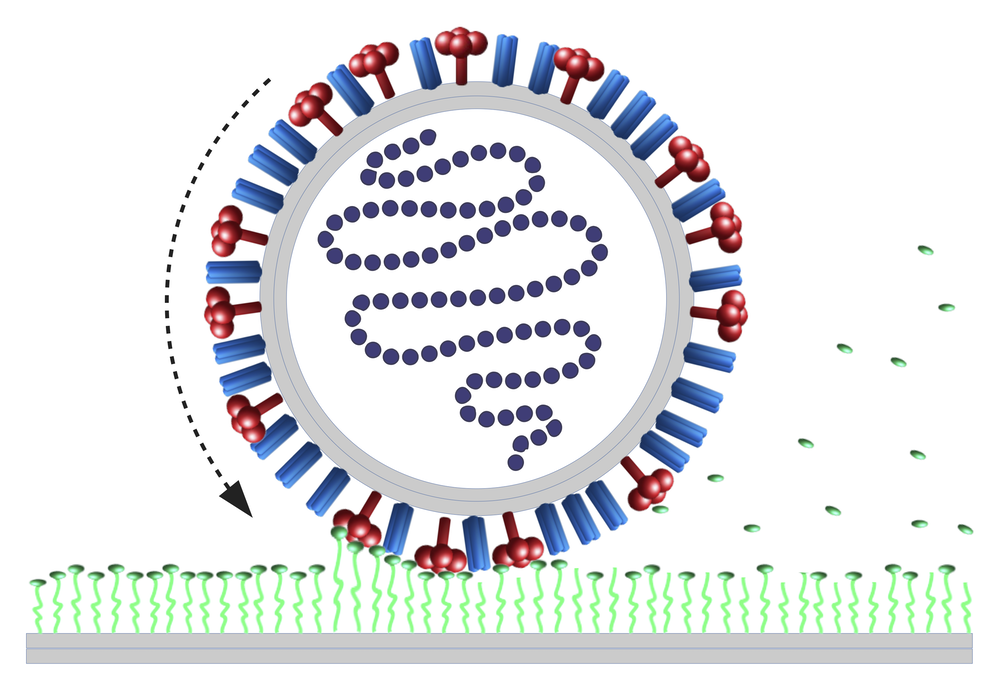
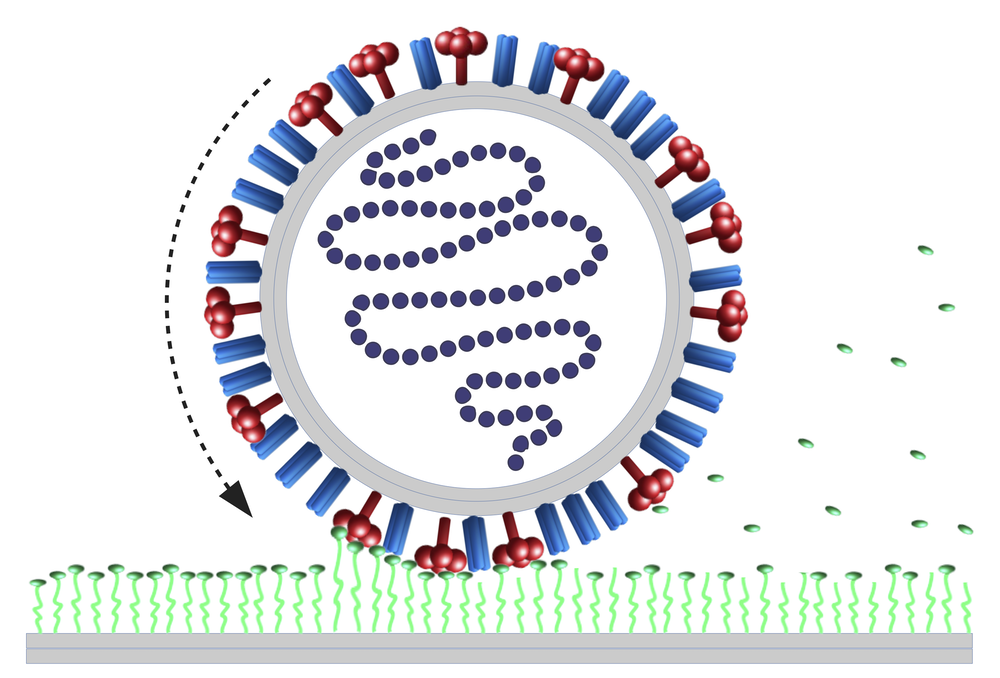
The tight grip is supplied by a “spike protein” called hemagglutinin (HA) that protrudes from the viral surface and binds to a chemical group called sialic acid on the surface of a cell membrane. The bond is temporary, as each HA continually disconnects and reconnects to a new sialic acid. Even so, with many of these bonds existing at any given time, they would hold the virus in place. But there is a second spike protein called neuraminidase (NA) that cuts off sialic acid groups in the contact region, gradually reducing the number of binding sites available for HA. Experiments show that without HA, the virus can’t attach; without NA, it can’t move [2].
But how do HA and NA act together to produce concerted rolling motion, rather than, say, a disorganized succession of binding and cleavage events? To explain that, physicists Falko Ziebert of Heidelberg University in Germany and Igor Kulić of the Charles Sadron Institute in Strasbourg, France, have now devised a model that adapts ideas developed for other biological “motors” that produce molecular-scale motion. They conclude that the virus rolling operates in a way not previously identified in biological systems.
The theorists model the influenza A virus (IVA) as a long cylinder studded with a random mixture of HA and NA spike proteins. First, the virus sticks to the membrane via several hundred HA–sialic acid links, and then the NAs gradually reduce the virus’s sticking strength by cutting off sialic acid groups. With the virus-cell connection weakened, the virus can wobble slightly around its attachment point, increasing the chances that some HAs just beyond the edge of the contact region form fresh links, pulling the cylinder into rotation.
Using both model calculations and computer simulations, the two researchers find that this process can produce persistent rolling motion. Once the symmetry of the system is broken, pulling the virus in one specific direction, the interplay of HA and NA creates a torque that keeps the virus rolling. Occasionally, the virus particles can, through chance fluctuations in the binding and cleavage events, reverse direction and set off in a new direction.
“The really surprising thing is that the rolling is almost independent of the system parameters,” such as the NA cleavage rate, says Kulić—it seems to be virtually inevitable as long as the virus remains stuck to the membrane. The researchers say that their proposed mechanism is probably similar to that operating in a recently reported artificial system involving silica nanoparticles coated with DNA that roll over a surface covered with RNA when a bond-cutting enzyme is present [3].
“If this result is correct, it would open important vistas for constructing very simple enzyme-powered molecular machines,” says biophysicist Dean Astumian of the University of Maine, a specialist in molecular and nanoscale motors. However, in the absence of a more detailed model, he is skeptical that the mechanism explains how the virus can break the symmetry and begin rolling steadily in one direction. Kulić agrees that the virus is frozen in place at first but says that that state doesn’t last.
Virologist Erik de Vries of Utrecht University in the Netherlands says that “any physical model or theory to dissect and quantify the rolling behavior of IVA is highly valuable.” But he says that more data on the reaction rates of HA and NA are needed to properly test the model.
Kulić points out that flu drugs such as Tamiflu work by inhibiting NA and thus blocking the rolling motion, immobilizing the virus until it can be flushed away. Some coronaviruses also move on cell surfaces, and versions of the HA and NA proteins are known to be involved, he says, but luckily SARS-CoV-2 does not move. “For now, SARS-CoV-2 is a one-trick pony, not well adapted to the host,” he says. “Let’s hope it does not figure out influenza’s rolling trick soon.”
–Philip Ball
Philip Ball is a freelance science writer in London. His latest book is How Life Works (Picador, 2024).
References
- F. Ziebert and I. M. Kulić, “How influenza’s spike motor works,” Phys. Rev. Lett. 126, 218101 (2021).
- E. de Vries et al., “Influenza A virus hemagglutinin–neuraminidase–receptor balance: Preserving virus motility,” Trends Microbiol. 28, 57 (2020).
- K. Yehl et al., “High-speed DNA-based rolling motors powered by RNase H,” Nat. Nanotechnol. 11, 184 (2015).