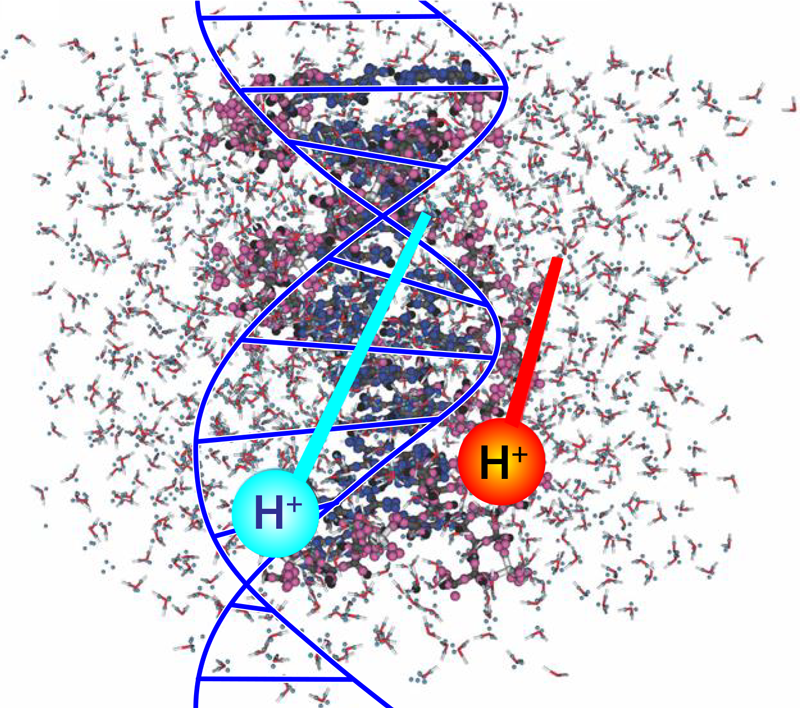
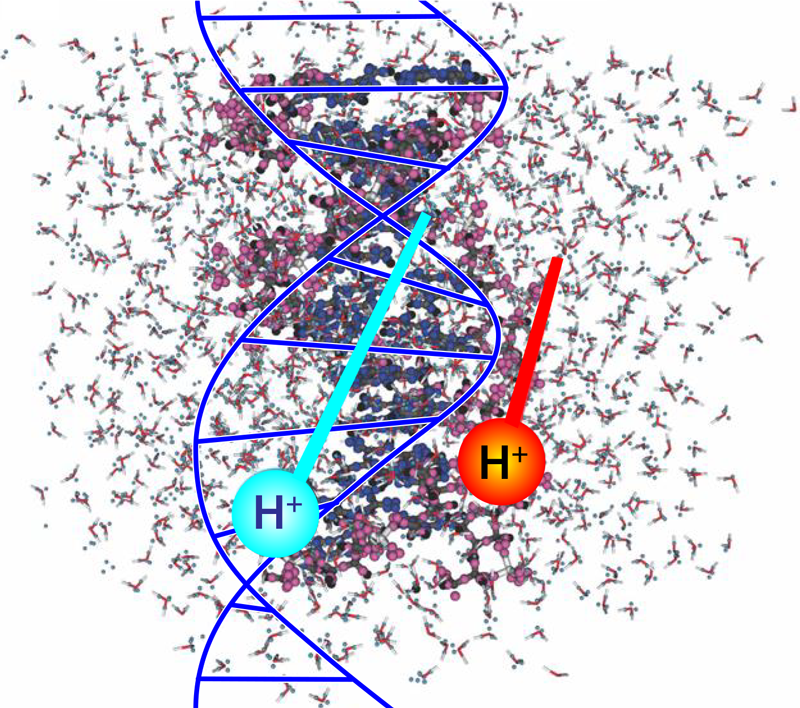
The Impact of Ions on DNA
Radiobiology studies on the effects of ionizing radiation on human health focus on the deoxyribonucleic acid (DNA) molecule as the primary target for deleterious outcomes. The interaction of ionizing radiation with tissue and organs can lead to localized energy deposition large enough to instigate double strand breaks in DNA, which can lead to mutations, chromosomal aberrations, and changes in gene expression. Understanding the mechanisms behind these interactions is critical for developing radiation therapies and improving radiation protection strategies. Christopher Shepard of the University of North Carolina at Chapel Hill and his colleagues now use powerful computer simulations to show exactly what part of the DNA molecule receives damaging levels of energy when exposed to charged-particle radiation (Fig. 1) [1]. Their findings could eventually help to minimize the long-term radiation effects from cancer treatments and human spaceflight.
The interaction of radiation with DNA’s electronic structure is a complex process [2, 3]. The numerical models currently used in radiobiology and clinical radiotherapy do not capture the detailed dynamics of these interactions at the atomic level. Rather, these models use geometric cross-sections to predict whether a particle of radiation, such as a photon or an ion, crossing the cell volume will transfer sufficient energy to cause a break in one or both of the DNA strands [4–6]. The models do not describe the atomic-level interactions but simply provide the probability that some dose of radiation will cause a population of cells to lose their ability to reproduce.
Because of its cell-neutralizing capability, ionizing radiation can be used to counter the growth of tumors. In fact, radiotherapy remains one of the most widely used cancer therapies [7, 8]. But when applied to treat malignancies, the therapy can also lead to severe outcomes for healthy tissues. In the case of gamma-ray and x-ray therapies, high-energy photons begin losing energy shortly after entering the body. In contrast, heavy-ion radiotherapy uses charged particles that lose most of their energy at the end of their travel range. Particularly for fast-moving particles, this rapid loss of energy over a very small distance leads to a sharp rise in the energy deposited in a localized volume. Because of this localized energy deposition, radiotherapists can use a charged particle beam to precisely target a tumor shape and depth, thus sparing healthy tissue in front of the tumor while minimizing damage to healthy tissues beyond the tumor. This selectivity makes heavy-ion radiotherapy a revolutionary therapeutic modality that may treat tumors that are traditionally considered incurable with current standard treatments.
Most of the energy transferred by a charged particle to a medium is the result of Coulomb interactions between electron orbitals. The average energy required to ionize an atom or molecule in a medium is often used to describe what’s known as the radiation stopping power of a material: the material’s ability to slow or stop charged particles, such as electrons or ions, as they pass through it [9]. Measuring a material’s stopping power is key to determining the utility of a radiation therapy. For biological tissues, stopping power is usually measured in terms of the energy lost per micrometer traveled. However, a DNA molecule has an average width of 2 nm, so measuring the stopping power at the scale of DNA isn’t currently possible.
Shepard and his colleagues used large-scale computational simulation on supercomputers to quantify the energy transfer from high-energy protons to solvated DNA, meaning a solution of DNA that’s separated into its sugar-phosphate side chains and nucleobase backbone components. They used time-dependent density-functional theory (DFT) to evaluate the DNA system’s complexity at the molecular level. DFT is a computational method to study the electronic structure of atoms, molecules, and solids. It is based on the concept that the properties of a many-electron system can be determined by a single function that describes the electron density of the system. DFT is an efficient method for calculating the electronic structure of large systems because it uses a set of approximations to account for the interactions between the electrons rather than solving the Schrödinger equation for each electron in the system. These approximations make it possible to calculate the electronic structure of complex systems that would be impossible to study using traditional methods.
In their simulations, the researchers expressed the total energy of the solvated DNA system as a mathematical function of the electron density. The electron density can be calculated from the system’s wave function, which, in turn, describes the probability of finding an electron in a particular position and with a particular spin. Using this approach, they found that the electron displacement was highly localized along the proton’s path and significantly higher in trajectories closer to the phosphate chains. The higher displacement indicates that the DNA’s sugar-phosphate backbone absorbed more energy than did the nucleobases.
The simulations call into question the conventional assumption that the stopping power is proportional to the number density of holes generated in the medium. Based on their results, Shepard and colleagues argue that the stopping power of the solvated DNA medium also depends on the energies of the generated holes. Their results indicated a higher prevalence of electron-hole formation in the sugar-phosphate backbone, which can lead to the formation of highly damaging free radicals. Free radicals are aqueous atoms or molecules that have an unpaired valence electron, making them highly reactive with the local medium. Radicals interacting with the sugar-phosphate backbone may cause fractures in the backbone and eventual breakage of one or more DNA strands.
This work demonstrates the utility and power of high-performance, multicore computers for studying complex interaction dynamics that are otherwise difficult to replicate in a laboratory setting. The results pinpoint where charged particles deposit most of their energy in a DNA molecule, helping bridge the gap in our knowledge of how radiobiology intersects with the physics of charged-particle transport. But some caution should be exercised in accepting the study’s conclusions until detailed experimental results validate the researchers’ models. With further clarification of the underlying mechanisms of DNA damage, scientists may be able to enhance the efficiency of therapeutic ionizing radiation. They may also be able to develop countermeasures, such as new drugs, that minimize the adverse effects of ionizing radiation on healthy cells.
References
- C. Shepard et al., “Electronic excitation response of DNA to high-energy proton radiation in water,” Phys. Rev. Lett. 130, 118401 (2023).
- J. Chancellor et al., “Everything you wanted to know about space radiation but were afraid to ask,” J. Environ. Sci. Health, Part C 39, 113 (2021).
- R. J. Preston et al., “Uncertainties in estimating health risks associated with exposure to ionising radiation,” J. Radiol. Prot. 33, 573 (2013).
- K. Chadwick et al., “The Molecular Model for Cell Survival Following Radiation,” in The Molecular Theory of Radiation Biology (Springer, Berlin-Heidelberg, 1981), p. 25[Amazon][WorldCat].
- C. A. Tobias, The Repair-Misrepair Model of Cell Survival (1980).
- S. B. Curtis, “Lethal and potentially lethal lesions Induced by radiation—A unified repair model,” Radiat. Res. 106, 252 (1986).
- J. Tobias, “The role of radiotherapy in the management of cancer–An overview,” Ann. Acad. Med. Singap. 25, 371 (1996).
- G. Delaney et al., “The role of radiotherapy in cancer treatment,” Cancer 104, 1129 (2005).
- J. F. Ziegler, “Stopping of energetic light ions in elemental matter,” J. Appl. Phys. 85, 1249 (1999).