The Mechanical Struggle behind Adaptive Immunity
To fight disease, many organisms have an adaptive immune system, which learns the molecular shapes of foreign elements (antigens) and remembers them to mount a defense against future infections. In vertebrates, the learning stage involves a remarkable cycle of evolution within an individual animal—a cycle called affinity maturation, which involves a type of immune cell called a B cell (Fig. 1). In this process, B cells are selected to have receptors that bind strongly to specific antigens. However, if these cells become too specialized, they risk becoming unresponsive to slightly mutated pathogens. Fortunately, the immune system can limit affinity maturation to retain a range of specificities for target pathogens. Just how the immune system is able to do that is the subject of a fascinating new study by Hongda Jiang and Shenshen Wang from the University of California, Los Angeles [1]. Their model incorporates a “struggle” between molecular forces that introduce variability in B-cell affinity maturation. It also provides a framework for addressing the trade-off between generalist and specialist immune strategies.
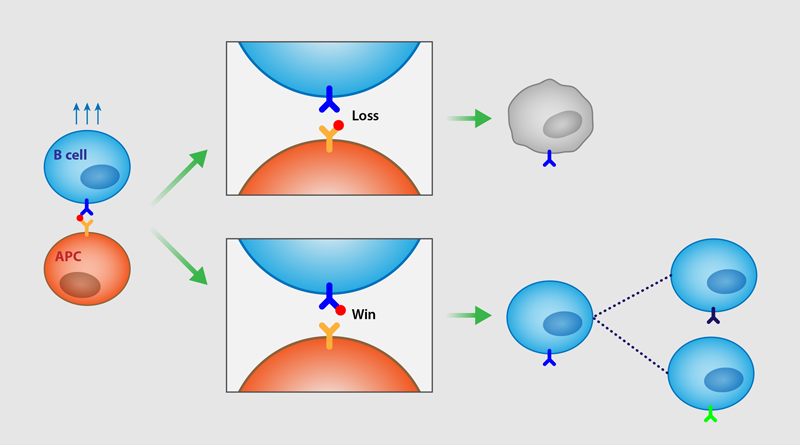
During affinity maturation, B cells collect within germinal centers—temporary structures within the lymph system where the B cells divide and undergo “somatic hypermutation.” In this reproduction step, the B-cell offspring present antigen receptors that vary slightly from those of their parent cells. For each generation, the B cells attempt to bind to and remove foreign elements from the surface of antigen-presenting cells (APCs) within the germinal center. Failure of extraction leads to programmed cell death (apoptosis), while success leads to the next cycle of affinity maturation (or, in some cases, the successful B cells differentiate into memory or plasma cells that exit the germinal center). In this way, the selection of B cells occurs through a rapid evolutionary process for more effective antigen recognition.
Jiang and Wang start from the observation that the survival and proliferation of a B cell within a germinal center depend on its ability to physically extract and internalize antigens presented by APCs. This process involves a molecular tug-of-war. Clusters of receptors on the B cell bind to antigen molecules, triggering intracellular signals directing the cell’s internal scaffolding to exert a pulling force. Meanwhile a tethering protein binds the antigen to the APC.
The researchers use analytical calculations and simulations to show how the length and stiffness of the tether balance with the binding affinity of the B-cell receptor to determine the likelihood of successful extraction of the antigen. The stiffer the tether, the stronger the receptor affinity needs to be to extract antigen and hence allow survival of the B cells. This creates a selective pressure driving evolution of the receptor in successive B-cell generations in the affinity maturation cycle. But once the receptor reaches a sufficiently strong affinity, the B cell generally wins the tug-of-war and so no selective pressure remains to keep tuning this antigen receptor. In sum, the researchers demonstrate a striking net effect: the intrinsic strength of the antigen tether sets an upper bound on the receptor binding strength achievable via affinity maturation.
Jiang and Wang also explore the possible mechanisms behind the diverse binding strengths found in B cells. They show that noise in molecular reaction networks (such as statistical variations in the concentrations of reagents or reaction products) can produce nongenetic variability in the force that B cells generate. This variability, combined with random differences in the physical properties of antigen tethers in the APCs, may explain why the maturation of a B-cell population leads to more than one type of receptor for a given antigen. This noise-based explanation raises a question: Is B-cell diversity simply a product of random molecular inputs, or has nature used that randomness to create an immune system that is better adapted to the pathogenic environment?
Recent studies have considered factors that should govern the organization of well-adapted immune systems. Some of this work has shown that the distribution of receptors should be skewed to prioritize rare threats—in other words, to prepare for uncommon pathogens that risk surprising the immune system [2]. Other work has discussed how diverse immune strategies employed by organisms ranging from vertebrates to bacteria could represent adaptations to different pathogen statistics [3]. Specific research on the CRISPR-based immune system of bacteria has found that the size of the immune repertoire may be limited by the number of special proteins used to read the stored immune memory [4, 5]. A key consideration in these previous studies was identifying the adaptive immune system’s principal line of defense against an evolving virus. Does it navigate the short-term dynamics of mutation, or does it rely on a long-term memory of the broad pathogenic landscape? As of now, the debate continues.
Ongoing work looks at constraints that may influence the level of diversity. For example, an organism needs to avoid having its immune system attack its own cells, and this autoimmunity constraint may set limits of cross-reactivity of vertebrate immune receptors [6, 7] and may control the size of DNA elements in the immune repertoire of bacteria [8]. In a similar vein, recent work considers affinity maturation in terms of the trade-off between short-term constraints and long-term immunity [9].
Against this context, the tug-of-war mechanism in Jiang and Wang’s study provides a new mechanism for maintaining an immune diversity that can respond to evolving pathogen landscapes while also mounting strong responses to current threats [1]. It will be interesting to see whether researchers will be able to use this insight to guide vaccine production or immunization protocol development, perhaps even by introducing bioengineered APCs that evince specific mechanical properties affecting affinity maturation.
References
- H. Jiang and S. Wang, “Molecular tug of war reveals adaptive potential of an immune cell repertoire,” Phys. Rev. X 13, 021022 (2023).
- A. Mayer et al., “How a well-adapted immune system is organized,” Proc. Natl. Acad. Sci. U.S.A. 112, 5950 (2015).
- A. Mayer et al., “Diversity of immune strategies explained by adaptation to pathogen statistics,” Proc. Natl. Acad. Sci. U.S.A. 113, 8630 (2016).
- S. Bradde et al., “The size of the immune repertoire of bacteria,” Proc. Natl. Acad. Sci. U.S.A. 117, 5144 (2020).
- A. Martynov et al., “Optimal number of spacers in CRISPR arrays,” PLoS Comput. Biol. 13, e1005891 (2017).
- J. K. Percus et al., “Predicting the size of the T-cell receptor and antibody combining region from consideration of efficient self-nonself discrimination.,” Proc. Natl. Acad. Sci. U.S.A. 90, 1691 (1993).
- C. J. E. Metcalf et al., “Demographically framing trade-offs between sensitivity and specificity illuminates selection on immunity,” Nat. Ecol. Evol. 1, 1766 (2017).
- H. Chen et al., “A scaling law in CRISPR repertoire sizes arises from the avoidance of autoimmunity,” Curr. Biol. 32, 2897 (2022).
- V. Chardès et al., “Affinity maturation for an optimal balance between long-term immune coverage and short-term resource constraints,” Proc. Natl. Acad. Sci. U.S.A. 119 (2022).