Protein Folding Can Be Surprisingly Slow
Understanding protein folding remains a fundamental unsolved problem in science. The basic question is, How does a protein chain consisting of perhaps thousands of linearly linked amino acids—20 varieties, each with a different chemistry, size, and shape—self-pack into a precise conformation? The structure of this conformation, the “native” state, is determined by the sequence in which the amino acids (called amino-acid “residues” when bound in a protein) have been arranged by evolution. While machine-learning algorithms have become expert in predicting what structure a particular protein sequence will fold to, computers—and humans—understand only poorly how and why that structure is achieved during folding. To shed light on these questions, researchers have tried to characterize the intermediate conformations adopted by a protein. Now a team led by Robert Tycko at the National Institutes of Health in Maryland has found evidence for an intermediate conformation that has not previously been seen in protein folding experiments [1]. The result indicates that the folding process takes much longer to complete than earlier studies had suggested.
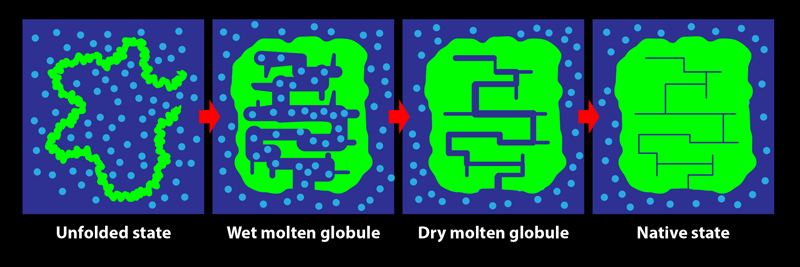
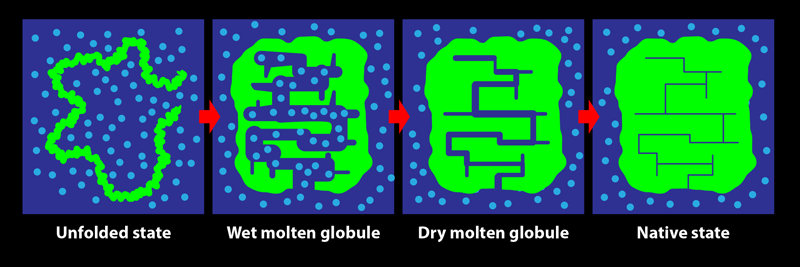
In its biological setting, a protein is surrounded by water molecules. During folding, the protein chain turns, twists, coils, loops, bends, and condenses, squeezing out water from within its structure. The early steps in this process are relatively accessible to experimental characterization by spectroscopic measures such as fluorescence and far-UV circular dichroism, which are sensitive to changes in the amount of water interacting with the chain. Tycko and his colleagues show that these investigative methods do not, however, reliably indicate the end of the folding process: consolidation to the final structure of a protein may proceed for a long time after the optical spectroscopic signals accompanying folding are complete. The team’s key result is the demonstration of the existence of a transient protein conformation called the dry molten globule (DMG) state.
The DMG was proposed in 1989 as the penultimate state adopted by a protein during folding [2]. It was envisaged as a slightly less compact form of the native state, in which internal contacts between parts of the protein are present, but in which the structure is not fully ordered and close packed. As the name suggests, the interior of the DMG-state protein was thought to be devoid of water. At the time, there was no experimental evidence for the DMG, and only the wet molten globule state (an expanded, hydrated, and fluctuating form with few long-range intrachain contacts) was known to exist.
Since that seminal theoretical study, experimental evidence for the state has come not from investigations of protein folding but from studies of the opposite process, with the formation of the DMG seen to be the first step during the unfolding of several proteins [3–6]. The initial expansion and partial disordering of the native protein structure to form the DMG was detected by energy-transfer methodologies, by various optical techniques, and by fluorine-19 nuclear magnetic resonance (NMR) measurements. The dryness of the DMG’s interior, meanwhile, was evident from the lack of water-dependent interactions with certain molecular probes and from the inability of NMR or mass-spectrometry-detected hydrogen-exchange measurements to detect the access of water to the buried main chain of the protein. In folding studies, fluorine-19 NMR experiments showed that residues may adopt their final, stable states at late stages of the process but were unable to detect the DMG state [7]. Although the formation of a DMG in the last but one step of folding, after water is extruded, had not been conclusively detected, a phosphorescence study of the folding of an enzyme provided hints [8]. That study indicated that the fold of the protein may continue to rigidify for days after a fully functional structure is achieved—as would happen during the DMG-to-native-state transition.
Tycko and his colleagues investigated a small protein consisting of 35 amino-acid residues—the “villin headpiece subdomain,” or HP35 (Fig. 1), known to be one of the fastest folding proteins. This is a three-helix protein that has been used extensively as a model protein for folding studies [9]. Optical spectroscopic methods have shown that the protein folds on the 10-µs timescale via multiple sequences of intermediate forms [10]. Now Tycko’s team has used solid-state NMR to show unequivocally that ordering and close packing of some residues occur well after the basic native fold is achieved.
In their experiment, they unfolded the protein using heat then refolded it at 30 °C. Refolding completes within 100 µs, according to optical spectroscopic probes. They kept the protein at 30 °C for an “incubation period,” which was varied from 1 to 10 ms, and then flash froze it. They characterized the frozen protein samples using carbon-13 solid-state NMR.
In protein NMR, the resonance for a certain atomic nucleus in a residue can become sharper when the molecular structure becomes more rigid. The team observed that the resonance frequencies of carbon-13 atoms in two specific residues, belonging to helices 1 and 3, did not change during the incubation period. This consistency indicated that these helices had already formed—that is, the protein had folded—by the start of the incubation period. On the other hand, resonances arising from several other residues spread over the protein sequence were seen to sharpen over the 10-ms incubation period. These results demonstrate that while the protein appears folded at the beginning of the incubation period, it actually becomes fully folded only at the end of it, with the residues losing their disorder and becoming rigidified in their final, well-packed conformations. The protein adopts the DMG state by 100 µs of folding then takes another 10 ms to reach its final, native conformation. It is not yet known how much greater stability is achieved during the DMG-to-native-state transition nor why the structural annealing takes so long. Nevertheless, this work [1] is the first definitive evidence for the formation of the DMG during folding.
It is yet to be established whether the DMG is a universal intermediate state in protein folding reactions. Tycko and his colleagues’ result [1] is a strong indication that it might be. It is commonly assumed that protein folding reactions are complete when there is no further change in the optical spectroscopic signals typically used to monitor them. This assumption is now shown to be wrong for HP35. If it turns out to be generally true that optical spectroscopic changes report on the formation of a DMG and not the native state of the protein, then many ideas about how proteins fold, such as the two-state (unfolded–folded) model and the mechanistic insights based on it, could be called into question [3, 4, 11]. The existence of a DMG would indicate that some and perhaps all of the intrachain contacts that form before the formation of the DMG are not tight, as usually assumed, but are, instead, liquid-like. The observation that close packing is achieved only when the DMG transits to the native state in the last step of folding points to the importance of van der Waals interactions in maintaining the integrity of native protein structure.
References
- C. B. Wilson et al., “Experimental evidence for millisecond-timescale structural evolution following the microsecond-timescale folding of a small protein,” Phys. Rev. Lett. 132, 048402 (2024).
- E. I. Shakhnovich and A. V. Finkelstein, “Theory of cooperative transitions in protein molecules. I. Why denaturation of globular protein is a first-order phase transition,” Biopolymers 28, 1667 (1989).
- S. K. Jha and J. B. Udgaonkar, “Direct evidence for a dry molten globule intermediate during the unfolding of a small protein,” Proc. Natl. Acad. Sci. U.S.A. 106, 12289 (2009).
- R. L. Baldwin et al., “Dry molten globule intermediates and the mechanism of protein unfolding,” Proteins: Struct., Funct., Bioinf. 78, 2725 (2010).
- S. Sarkar et al., “Unfolding of a small protein proceeds via dry and wet globules and a solvated transition state,” Biophys. J. 105, 2392 (2013).
- S. Neumaier and T. Kiefhaber, “Redefining the dry molten globule state of proteins,” J. Mol. Biol. 426, 2520 (2014).
- C. Frieden, “The kinetics of side chain stabilization during protein folding,” Biochemistry 42, 12439 (2003).
- V. Subramaniam et al., “Phosphorescence reveals a continued slow annealing of the protein core following reactivation of Escherichia coli alkaline phosphatase,” Biochemistry 34, 1133 (1995).
- J. Kubelka et al., “Chemical, physical, and theoretical kinetics of an ultrafast folding protein,” Proc. Natl. Acad. Sci. U.S.A. 105, 18655 (2008).
- S. Nagarajan et al., “Heterogeneity in the folding of villin headpiece subdomain HP36,” J. Phys. Chem. B 122, 11640 (2018).
- R. L. Baldwin and G. D Rose, “Molten globules, entropy-driven conformational change and protein folding,” Curr. Opin. Struct. Biol. 23, 4 (2013).