Molecular Lawnmower Drives Itself
Imagine a lawnmower that gets its driving directions from the grass that it is cutting. Biophysicist Nancy Forde of Simon Fraser University, Canada, and her colleagues not only imagined such a device, but they built a molecule-scale one out of proteins and a microsphere [1]. Driven by thermal fluctuations, the motor moves around a “lawn” of protein fragments, cleaving these grass blades as it goes and steering toward the next uncut patch.
Their design is an example of a molecular motor. In nature, molecular motors are protein-based structures that power directional motion at the cellular level. By converting chemical energy into mechanical energy, these natural motors facilitate cell locomotion and division, cargo delivery, tissue maintenance, and other processes fundamental to life. Inspired by these efficient machines, bioengineers have designed artificial versions using synthetic molecules, which typically achieve directional motion in the form of one-way rotation. But creating a molecular motor capable of directional motion from nature’s own building blocks has remained elusive.
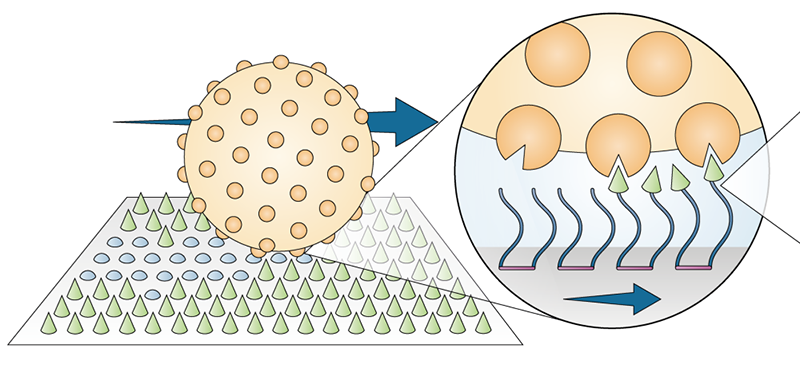
Forde and her colleagues based their lawnmower design around an enzyme called trypsin that aids with digestion. In our bodies, trypsin’s role is to break bonds between amino acids in proteins. Trypsin’s cutting machinery—which the team envisions as a biting “Pac-Man”—can be used to transform the chemical landscape that surrounds a molecular motor.
To build their motor, the researchers chemically attached thousands of Pac-Man-like trypsin enzymes to a 3-µm-diameter polystyrene-based bead. The decked-out motor looks like a “hairy bead,” says Chapin Korosec, the graduate student who led the work and is currently at York University, Canada. The team placed the motor onto a surface consisting of millions of short protein fragments called peptides bound to a silica substrate. Under a microscope, the researchers tracked the motor’s movements on the peptide lawn for 12.5 hours. They also performed a control experiment where the motor was set loose on a bare, peptide-free surface.
The data showed a distinct difference between the motion on the two surfaces. The motor performed a random walk on the bare surface, similar to a particle diffusing through a liquid. By contrast, the motor moved in quick, directional steps on the lawn, leaving patches of snipped peptides in its wake. On a grassy field, the lawnmower traveled at 58 nm/s, which is more than twice as fast as the diffusive motion of the motor on the bare surface—and slightly faster than other similarly sized synthetic molecular motors. In another experiment, the researchers patterned the lawn with microfabricated ridges. The lawnmower readily followed the guided tracks and moved even faster.
The lawnmower’s operating mechanism is known as a burnt-bridge Brownian ratchet. Driven by thermal energy, the motor will tumble in a random direction, but it has a preference to move toward uncut lawn.“Trypsin has a stronger binding affinity to intact peptide than to cleaved peptide,” explains Forde. If the motor lands in a region of uncut peptide, its trypsins will get to work, cleaving the newly reached lawn and creating a local asymmetry on the surface. In this way, “it’s ratcheted away from the burnt part of the lawn, toward the unvisited region,” explains Forde.
Ryota Iino, who designs molecular machines at the Institute for Molecular Science in Japan, called the study a “landmark paper” that “opens the possibility of redesigning various enzymes into molecular motors.” Cécile Fradin, a biophysicist at McMaster University in Canada, says that using proteins designed to do one thing and reassembling them to do something different “really appeals to the child in all of us who used to play with Legos and do things like repurposing bricks supposed to be part of a roof to build a dragon.”
This repurposing of proteins is “truly a remarkable feat,” says Lukas Pfeifer, at the Swiss Federal Institute of Technology in Lausanne (EPFL), who developed a photon-driven molecular motor. One possible future direction, he says, is depositing motors on a substrate that has an inhomogeneous concentration of surface-bound peptides. The motors would naturally move toward regions with the highest local concentrations—a step toward targeted drug delivery. Another possibility would be to use different enzymes that could guide motors down different tracks—the starting point for a future biocomputer. “Since nature appears to prefer proteins as the functional motor elements, and proteins provide a vast diversity of function, it is exciting to be able to start exploring their potential as elements of our motor engineering toolbox,” says Forde.
–Rachel Berkowitz
Rachel Berkowitz is a Corresponding Editor for Physics Magazine based in Vancouver, Canada.
References
- C. S. Korosec et al., “Motility of an autonomous protein-based artificial motor that operates via a burnt-bridge principle,” Nat. Commun. 15, 1511 (2024).