X-Ray Imaging of a Single Virus in 3D
X-ray crystallography has been a defining tool for structural biology. Since the determination of the molecular structure of myoglobin in 1957, the technique has allowed researchers to determine the structure (and hence the function) of tens of thousands of proteins, nucleic acids, and other biological molecules. For the method to work, the molecules of interest have to be assembled into a crystal that can be investigated by monitoring how an x-ray beam scatters off of it. Unfortunately, many important molecules do not cooperate: they are impossible to crystallize. Addressing this limitation has been a key driver of a new generation of light sources based on x-ray free-electron lasers (XFELs). An international community of XFEL users is seeking to reconstruct molecular structure without crystals by scattering x rays off single molecules injected into powerful XFEL beams. A team lead by Janos Hajdu at Uppsala University, in Sweden, has now reported a key step towards this goal [1]. They have demonstrated that it is possible to reconstruct the three-dimensional structure of a virus from a very large number of diffraction patterns collected from a sequence of randomly oriented single viruses.
The Achilles’ heel of x-ray crystallography is captured in its name. Crystallography is able to provide exquisite and beautiful structures of complex molecules, but it can only do so if the molecules are arranged in a perfect and relatively large crystal. But many important molecules cannot be crystallized, such as proteins that interact with biological membranes (“membrane proteins”). These proteins play a role in the immune response, in cellular ion channels and receptors, and they are so crucial for life that a very large fraction ( – ) of the human genome is dedicated to keeping the code for their production. So, how can one determine the structure of a biomolecule that will not crystallize? This is where XFELs come in. These facilities produce highly coherent x-ray pulses that are some 10 orders of magnitude brighter than the beams of conventional synchrotrons. The pulse coherence allows the realization of diffractive imaging techniques—able to reconstruct a sample’s image by measuring its diffraction pattern. The hope is that XFELs can produce pulses so bright that scattering can be seen from a single biomolecule, and so short (a few femtoseconds) that scattering occurs before the molecule disintegrates under the intense x-ray pulse.
Even though XFEL beams are very intense, the scattering from a single molecule would be too weak to generate a signal large enough to yield the full molecular structure. Instead, experiments will have to add together the scattering patterns from many single molecules. Several key questions must be tackled before such experiments become a reality, but they are being progressively answered. First, are the x-ray pulses short enough to be able to diffract off the molecule and recover structural information before the molecule inevitably explodes? Theoretical and experimental results on this aspect are encouraging: Neutze et al. [2] modeled the behavior of biomolecules in an intense x-ray beam and concluded that pulses shorter than about 5 femtoseconds would be necessary. And Chapman et al. [3] have carried out a proof-of-principle experiment, imaging a nanopatterned structure (two stick men walking under the sun) before the sample exploded. Second, can one recover an image from the intensity of the field diffracted by a noncrystalline object? The field of coherent diffraction imaging—developed over the last 15 years since the original demonstration by Miao et al. [4]—has shown this is indeed possible. Finally, can one collect diffraction patterns from tens, hundreds, thousand, or even millions of individual molecules and then put together these noisy data sets, by aligning the measured diffraction patterns to compensate for the random orientation of the molecules? The work of Hajdu’s team demonstrates this key capability.
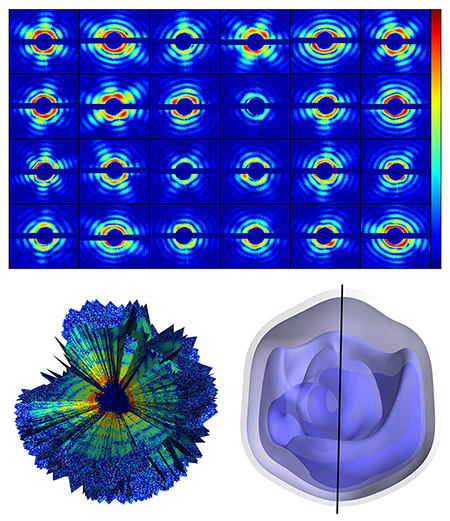
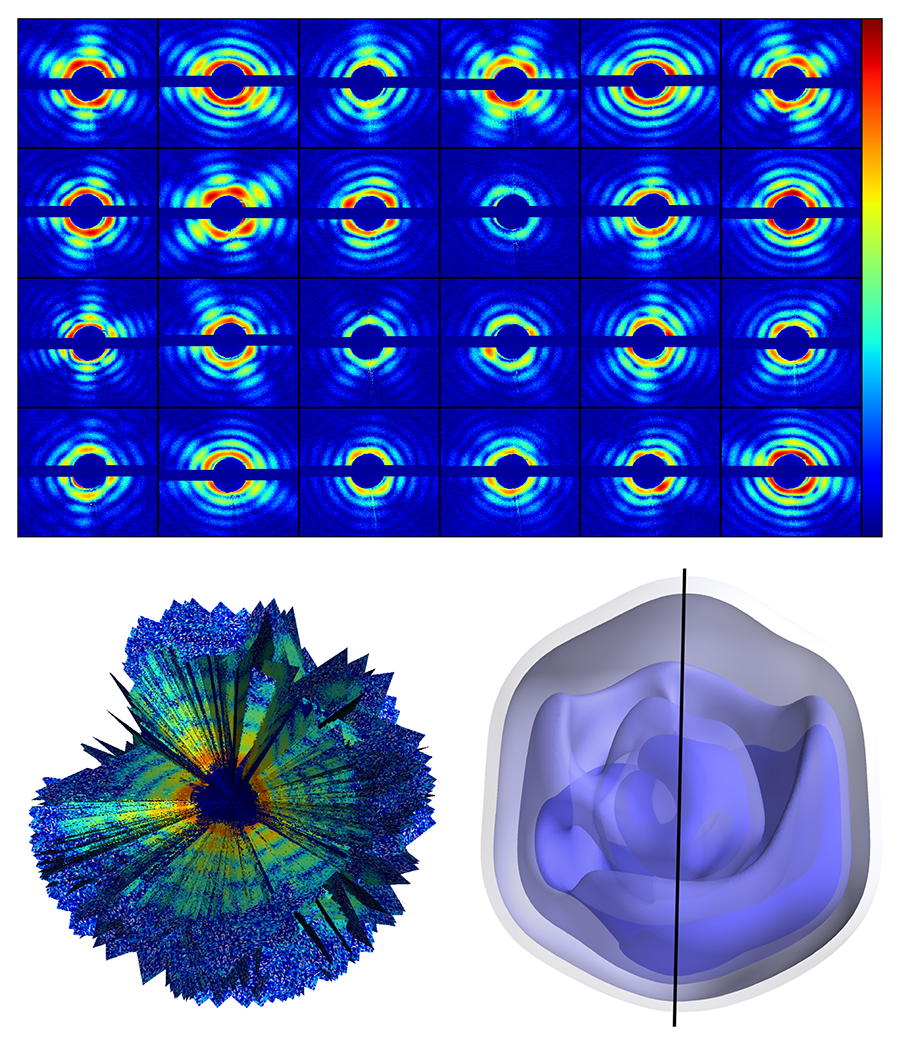
The authors used the giant mimivirus as their target—a simpler case than that of a small biomolecule. The mimivirus is one of the largest known viruses—about half a micrometer in diameter. First isolated in 1992 from amoebae growing in a water tower, it is thought to be the cause of certain forms of pneumonia. Compared to smaller proteins, such a large virus is relatively easy to inject into the x-ray beam, is more robust to changes in its environment, and scatters far more x rays than even the largest biomolecules. The team injected single viruses into an x-ray beam at the Linac Coherent Light Source at the Stanford Linear Accelerator Collider, in the US, the first operating hard x-ray free-electron laser and one of only two in the world (the other is the SACLA facility in Japan). The viruses were sprayed into the x-ray beam and a series of diffraction patterns were recorded. Because the sample is isolated and nonperiodic, the diffraction data is a diffuse pattern (see Fig. 1, top), without the discrete x-ray Bragg diffraction spots displayed by a crystalline structure. A total of 198 diffraction patterns were collected. The orientation of the virus for each diffraction pattern is randomly distributed and unknown.
To sort the set of diffraction patterns and assemble them into a three-dimensional diffraction dataset, the team exploited mathematical methods developed in 2009 at Cornell University by Veit Elser and Duane Loh [5] (see Fig. 1, bottom left). Through an iterative optimization algorithm, the method allows the recovery of the relative orientation of the particle captured by each snapshot, and, by adding up all contributions, the reconstruction of a complete 3D diffraction pattern (which represents the Fourier transform of the electron density of the sample). Then, through phase recovery methods previously developed for x-ray coherent diffractive imaging, the diffraction pattern can be inverted to obtain a three-dimensional image of the giant mimivirus.
The measurement yielded a virus image with a spatial resolution of 125 nanometers (see Fig. 1, bottom right), which is not in itself particularly impressive: The experiment is unlikely to reveal any striking new biological insights. But that is not the point. This team has demonstrated that it is possible to gather noisy diffraction data from hundreds of individual samples, figure out the relative orientation of all of them, and assemble the data into a complete and consistent three-dimensional diffraction pattern, from which the 3D structure can be derived.
The work of Hajdu and his colleagues thus represents a crucial step forward, completing the set of methods that, in principle, allow single-molecule images to be reconstructed. With large enough signals, we now know how to put together a series of single-particle data. Since we already know how to solve the phase problem for coherent diffraction data,, we can be reasonably confident that suitable data can be analyzed. But this does not mean that the imaging of small isolated biomolecules is around the corner: several obstacles must still be overcome. Researchers must test ways to reliably inject a single molecule, without damaging it or altering its structure, into the x-ray beam. And there are still open questions on the impact of electronic damage on x-ray scattering on femtosecond time scales: The above-mentioned work by Neutze et al. tracked the movements of the atomic nuclei of the biomolecule, showing they don’t move on the few-femtosecond timescale of an x-ray pulse. Electrons, however, move faster than nuclei. Since electrons are what scatters x rays, it is yet to be confirmed that few-femtosecond pulses can probe an unperturbed electronic structure. Finally, single-molecule experiments will have to be accompanied by a substantial improvement in the spatial resolution for the results to become biologically interesting. But progress on all these fronts is proceeding at a very rapid pace.
This research is published in Physical Review Letters.
References
- Tomas Ekeberg et al., “Three-Dimensional Reconstruction of the Giant Mimivirus Particle with an X-Ray Free-Electron Laser,” Phys. Rev. Lett. 114, 098102 (2015)
- R. Neutze, R. Wouts, D. van der Spoel, E. Weckert, and J. Hajdu, “Potential for Biomolecular Imaging with Femtosecond X-Ray Pulses,” Nature 406, 752 (2000)
- H. N. Chapman et al., “Femtosecond Diffractive Imaging with a Soft-X-Ray Free-Electron Laser,” Nature Phys. 2, 839 (2006)
- J. Miao, P. Charalambous, J. Kirz, and D. Sayre, “Extending the Methodology of X-Ray Crystallography to Allow Imaging of Micrometre-Sized Non-Crystalline Specimens,” Nature 400, 342 (1999)
- Ne-Te Duane Loh and Veit Elser, “Reconstruction Algorithm for Single-Particle Diffraction Imaging Experiments,” Phys. Rev. E 80, 026705 (2009)