How Cells Remember Who They Are
How does a liver cell remember that it's a liver cell when it replicates? A cell's identity is determined in part by small molecules that bind to chromosomes at specific locations. Now researchers have devised a model that seems to explain how these so-called epigenetic patterns are sustained across generations of cells. The model connects the epigenetic marking of genes with the three-dimensional structure of DNA as it is folded up inside chromosomes. It leads to a mechanism for patterns of markings to be repaired if they are disturbed by events such as gene replication.
Epigenetic markings are small molecules added onto parts of chromosomes that may act as genetic switches, silencing some genes and turning on others, so that cells become different types—liver, muscle, or heart cells, for example. Chromosomes are made of chromatin fibers, which are linear DNA molecules looped around cylindrical histone proteins, forming a "string of pearls." A genome’s epigenetic annotations also influence the way that chromatin folds into a three-dimensional shape. This complex folding also affects the activity of genes by preventing the transcription machinery from easily accessing DNA regions that are not at the surface. But the relationships among epigenetic marking, chromatin folding, and gene activity are not completely clear.
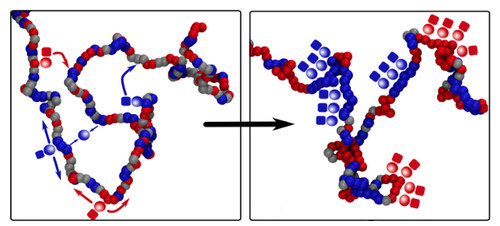
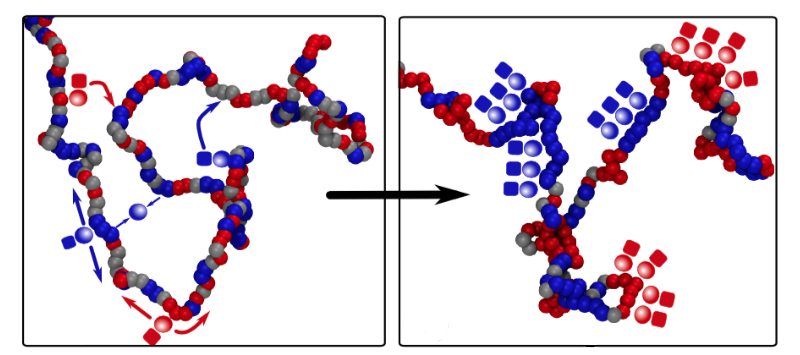
Davide Michieletto of the University of Edinburgh in the UK and his co-workers constructed a model of epigenetics that is much simpler than the real biological picture but retains the essential ingredients. They consider a chromatin fiber as a string of “beads,” each bead corresponding to about ten DNA-wrapped histones, and assume that each bead can be epigenetically marked in just two different states, represented by red and blue. (Beads may also be unmarked, represented by gray.) Markings of the same color attract one another, mimicking the way protein molecules that “read” the epigenetic instructions typically bridge several identical epigenetic markings at once, bringing those regions of DNA together temporarily. Markings of different colors, meanwhile, repel each other.
There is experimental evidence that in real cells the proteins that write epigenetic marks are recruited by proteins that read the same mark. This protein association can help to sustain epigenetic memory via a positive feedback loop: even if some marks get erased, the presence of others in the same region will encourage the erased marks to be written back again. Michieletto and colleagues mimic this process in their model by allowing beads to be recolored at a certain rate, with the color choice biased to favor the prevailing color in the region.
The researchers find that an initially random distribution of colors can switch to an ordered state in which the fiber is compactly folded, and the colors are the same over large domains or even the entire chromatin fiber. This change happens abruptly in a so-called first-order phase transition—like water to ice—as long as the bead attraction is strong enough. The ordered state is stable in the presence of occasional disruptions, just as ice will always re-form in the freezer, even if there is some partial melting when someone opens the door. The same basic behavior persists even when the model includes a more realistic description of the kinetics of epigenetic writing.
“First-order transitions are robust against perturbations,” Michieletto explains, “and so the system has memory.” So even if a large proportion of the epigenetic marks get scrambled, for example, as a cell replicates its genome in preparation to divide, the genome can find its way back to its previous state through a series of rewriting steps. “This aspect is very important for stability and cellular identity,” says Michieletto.
“I really think that this work has great potential,” says Sonja Prohaska, a specialist in epigenetics at the University of Leipzig. “I’m impressed how well the physics and the modeling embed a broad range of observations." She finds the work "astonishingly convincing, and [it] demonstrates that physics has a place in current biology and that it might even simplify a biologist’s life.”
Mario Nicodemi of the University of Naples in Italy says that it is unclear how the model can be tested by experiments. Michieletto admits that it may take some years before the necessary technology becomes available to make such tests but sees several possibilities. For example, one could manipulate a cell's genes to make it produce non-functional reading or writing proteins, breaking the positive feedback on which the epigenetic memory depends, and compare the effects with the model's predictions.
This research is published in Physical Review X.
–Philip Ball
Philip Ball is a freelance science writer in London. His latest book is How Life Works (Picador, 2024).