Getting a Handle on Proteins
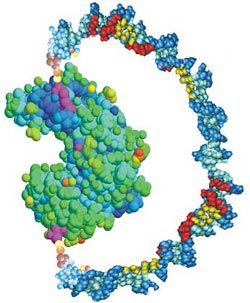
A U-shaped handle of DNA attached to an enzyme molecule can apply enough force to disrupt the enzyme’s activity, report biophysicists in the 12 August PRL. The researchers could control the enzyme’s effectiveness by stiffening the DNA, which pulled on the protein, like a bow pulling its string taut. This technique could help researchers understand how enzyme structure determines function and suggests a way of creating new types of drugs.
Enzymes are protein molecules that contain pockets for binding other molecules and catalyzing reactions on them. Many enzymes also have separate binding regions, called allosteric sites, where smaller molecules bind to alter the reaction rate of the enzyme, speeding it up or slowing it down. Researchers believe that binding to an allosteric site distorts the enzyme’s structure or changes its flexibility, which affects the fit between the enzyme and its target molecules, and therefore the reaction rate. But they don’t fully understand the complex intra-molecular events involved in allosteric control.
“The question is why you need such a complicated [protein] molecule to have this capability” of regulated activity, says Giovanni Zocchi of the University of California, Los Angeles (UCLA). To learn more about allosteric control, he and his colleagues have designed their own protein control technology. Some researchers have tried engineering allosteric sites that bind metal ions, but this strategy is relatively complicated and dependent on the details of the enzyme. “The uniqueness of our approach is that it can be applied to any protein,” says Zocchi.
In the strategy developed by Zocchi, graduate student Brian Choi, and their UCLA colleagues, a handle-like strand of DNA is attached to a protein using chemistry and molecular biology techniques. A single strand of DNA is floppy, so it has little effect on the protein. Add a complementary DNA strand into the solution, however, and the two condense into a more rigid double helix that wants to straighten out. The stiffer DNA pulls taut against the protein and exerts up to 10 piconewtons of force, the researchers estimate. They tested the method on a vise-shaped enzyme called guanylate kinase, whose job is to bind two small molecules, ATP and GMP, and move a phosphate ion from one to the other. The jaws of the enzyme close around its target molecules, and the team oriented the DNA so that the stiffer double helix would resist the jaws’ closure.
To determine the effect of the rigid, 20-nanometer-long double helix on the enzyme, the team used standard biochemical assays to measure the rates at which the enzyme binds to GMP and ATP. Its affinity for GMP fell by a factor of ten, while its affinity for ATP remained within a factor of two of that of the enzyme-plus-single-strand. They concluded that the tension applied by the double helix disrupted the GMP binding site but left the ATP binding site relatively unaffected. Eventually the method might be turned into a therapeutic tool, says Zocchi, in which the DNA-protein fusion is activated by a piece of DNA or RNA present only in diseased cells such as tumors.
“The authors achieved a direct mechanical influence on chemical processes,” says Elisha Moses of the Weizmann Institute of Science in Rehovot, Israel. “This is conceptually new and exciting.” The method might serve as a diagnostic tool to detect the presence of genes active in disease, says David Bensimon of the Ecole Normale Supérieure in Paris, providing it demonstrates high efficiency and sensitivity.
–JR Minkel
JR Minkel is a freelance science writer in New York City.


