A Gentle Twist on DNA
DNA is the storage medium for the genetic information in cellular life, but it is also increasingly used as a building block in bio- and nanotechnological applications. A fundamental property of DNA that is of critical importance in both cases is its torsional stiffness—its resistance to deforming under torsional stresses. This property, which derives from the molecule’s iconic right-handed double-helical structure, links processes such as replication, transcription, or DNA repair to rotational motion and torsional strains [1]. This stiffness also controls how torsional strain affects the molecule’s shape: beyond a certain threshold, torsion can cause the double helix to form higher-order structures called plectonemic supercoils, reminiscent of the inevitable tangles of an old-fashioned spiral telephone cord. Such DNA supercoiling regulates DNA readout and processing. Xiang Gao and his colleagues in the Michelle Wang lab at Cornell University, New York, have now measured the torsional stiffness of DNA—and of its tangled, plectonemic form—using a new variation on an ingenious single-molecule approach called an optical torque wrench [2]. The method could allow researchers to probe DNA behavior in the biologically relevant regime of low forces and constant extensions.
The first measurements of DNA torsional stiffness used DNA cyclization [3] and fluorescence polarization anisotropy [4], two indirect methods whose accuracy is limited by the assumptions of the models needed to interpret the data. Subsequently, single-molecule manipulation approaches led to more direct measurement methods, including a rotor bead assay [5], magnetic torque tweezers [6], and the optical torque wrench developed by Wang’s group [7]. These single-molecule methods confirmed a theoretical prediction delivered by the so-called Moroz-Nelson (MN) model: the effective torsional stiffness of DNA depends on how strongly the molecule is stretched while the torsional strain is applied [8]. Increasingly precise measurements, however, suggested discrepancies between experimental data and MN-model predictions in the physiologically relevant regimes where the stretching forces are small. These deviations helped to refine the theoretical models by, for example, taking into account the anisotropy of the DNA helix [9] or the possibility of transitions between different conformations of the DNA backbone [10].
An outstanding question involves the effect of additional DNA-molecule turns that increase the DNA linking number—the total number of windings of one DNA strand around the other. This increased winding is accommodated by either twist (local torsional strains of the DNA helix), writhe (the number of times the axis of the double helix crosses itself), or a combination of the two. Early work using electron microscopy suggested that when the linking number of unstretched, circular DNA is increased (requiring the circle to be broken, twisted, and then remade) % of the excess is accommodated as twist and % as writhe, independent of the absolute linking number [11]. In contrast, in measurements of single molecules under tension, two regimes emerge, depending on the excess linking number. At low excess linking number, virtually 100% of the excess is absorbed as twist. But for linking numbers beyond which a buckling transition occurs (when plectonemes start to form), the excess linking number is mostly absorbed as writhe. How to reconcile these seemingly disparate situations?
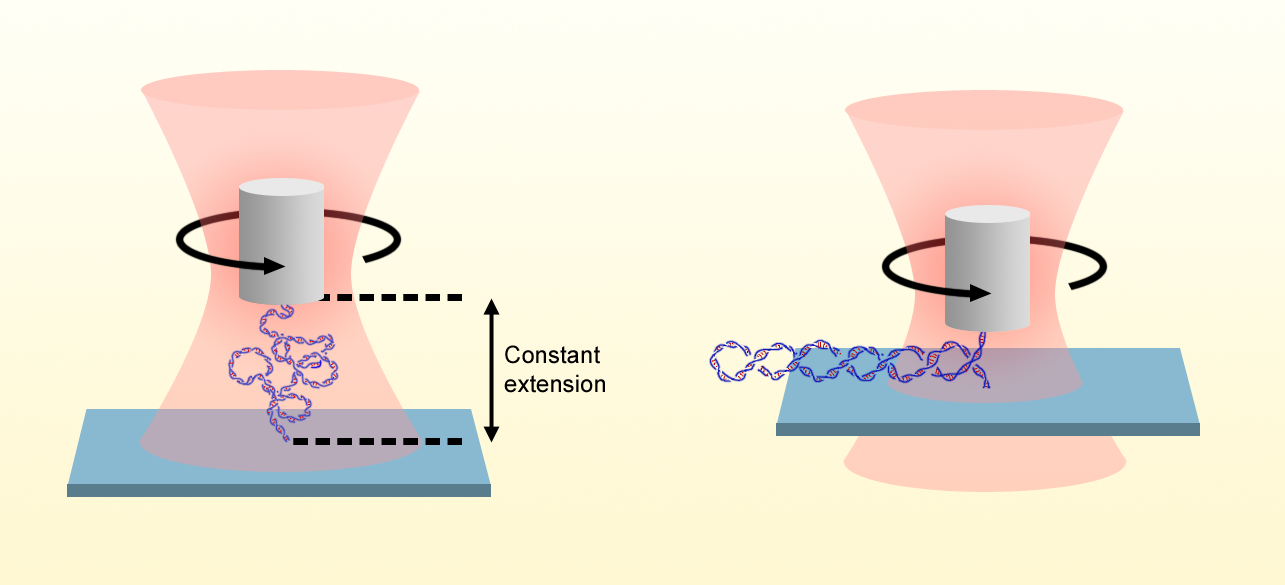
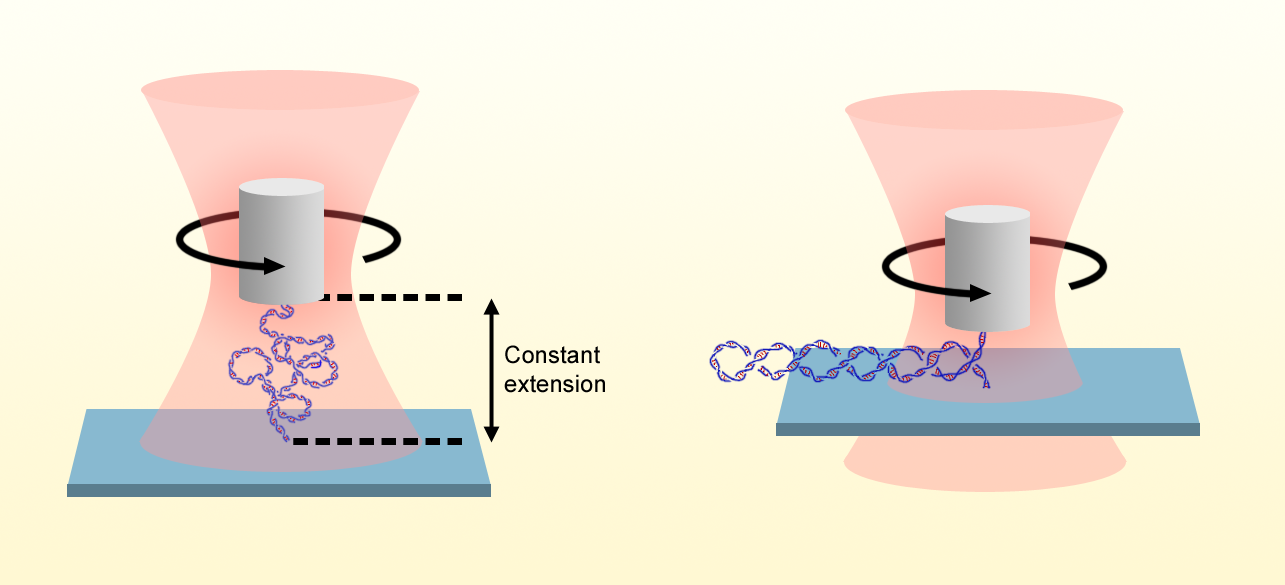
Gao and his colleagues address this question by building on their previously demonstrated optical torque wrench. In this method, a nanofabricated birefringent quartz cylinder is held in optical tweezers in which the polarization of the laser is controlled and measured. One end of a single DNA molecule is attached to the quartz cylinder, and the other end is attached to a stationary surface. By varying the height of the optical trap, the researchers stretch the DNA, and by rotating the input laser-beam’s polarization, they apply additional turns via rotation of the cylinder. They then determine the torque on the molecule by detecting the change in the angular momentum of the output beam after it has been transmitted through the cylinder.
Whereas previous torque measurements were made while the DNA was under constant tension, the researchers exploit the excellent spatiotemporal control of their setup to establish a new measurement mode, in which the extension of the molecule, rather than the applied force, is held constant. This feature is crucial because measurements of the extension can be made with a smaller error than measurements of the applied force, which may have an unknown offset. The approach allowed the Wang group to expand the available DNA torsional stiffness data down to forces of 20 fN—an order of magnitude smaller than the previous state of the art.
The new data show that the torsional “persistence length” , which measures the molecule’s torsional stiffness, approaches a value of nm under 20 fN of tension and clearly confirm deviations from the MN model at low forces. Gao and his colleagues also measure the torsional stiffness of the plectonemic state to be nm, in excellent agreement with indirect estimates from fitting a two-state model [12] to buckling torque data [6]. The similar values of and suggest that, in the low-force regime, excess twist is partitioned approximately equally between the straight and plectonemic phases upon addition of supercoils. Further, the values of and at the lowest forces indicate that torsional stiffness is % of that seen at high forces, suggesting that excess linking number partitions into 25% twist and 75% writhe. This result is similar to that of circular DNA [11], which implies that the partitioning of linking number is independent of the chain topology (linear vs circular).
These findings are interesting in their own right, but we envision that the constant-extension measurement mode demonstrated by the Wang group will allow ever more detailed studies of DNA and of its interaction with proteins within this hard-to-probe, small-force regime. Having access to this regime is exciting because of its relevance for biology, where forces are small. Furthermore, the constant DNA extension is consistent with the situation in the cell, in which DNA is confined in domains that are separated by hardly movable barriers. The methodology should also be widely applicable to other contexts and other macromolecules, such as DNA origami-type assemblies or double-stranded RNA.
References
- J. Lipfert et al., “Torque spectroscopy for the study of rotary motion in biological systems,” Chem. Phys. 115, 1449 (2014).
- X. Gao et al., “Torsional stiffness of extended and plectonemic DNA,” Phys. Rev. Lett. 127, 028101 (2021).
- D. Shore and R. L. Baldwin, “Energetics of DNA twisting,” J. Mol. Biol. 170, 957 (1983).
- B. S. Fujimoto and J. M. Schurr, “Dependence of the torsional rigidity of DNA on base composition,” Nature 344, 175 (1990).
- Z. Bryant et al., “Structural transitions and elasticity from torque measurements on DNA,” Nature 424, 338 (2003).
- J. Lipfert et al., “Magnetic torque tweezers: Measuring torsional stiffness in DNA and RecA-DNA filaments,” Nat. Methods 7, 977 (2010).
- S. Forth et al., “Abrupt buckling transition observed during the plectoneme formation of individual DNA molecules,” Phys. Rev. Lett. 100, 148301 (2008).
- J. D. Moroz and P. Nelson, “Torsional directed walks, entropic elasticity, and DNA twist stiffness,” Proc. Natl. Acad. Sci. U.S.A. 94, 14418 (1997).
- S. K. Nomidis et al., “Twist-bend coupling and the torsional response of double-stranded DNA,” Phys. Rev. Lett. 118, 217801 (2017).
- J. M. Schurr, “A quantitative model of a cooperative two-state equilibrium in DNA: Experimental tests, insights, and predictions,” Q. Rev. Biophys. 54, e5 (2021).
- T. C. Boles et al., “Structure of plectonemically supercoiled DNA,” J. Mol. Biol. 213, 931 (1990).
- J. F. Marko, “Torque and dynamics of linking number relaxation in stretched supercoiled DNA,” Phys. Rev. E 76, 021926 (2007).