Model of Chromosome Replication Gets Upgraded
An Escherichia coli bacterium takes about 40 minutes to replicate its chromosome. Replication is a necessary precursor to cell division, and yet Escherichia coli can divide every 20 minutes. How is this possible? The answer is that, before a round of replication is over, the bacterium has already initiated the next one (or two)—as biologists have known for more than 50 years. But deciphering how exactly replication is initiated has since remained an unresolved central problem in bacterial physiology. Now a theoretical study by Haochen Fu and colleagues from the University of California, San Diego, brings important new understanding to this problem [1]. They present a model that offers elegant explanations for two puzzling features of replication initiation: why the protein that initiates replication switches between two states even though only one is capable of initiation, and why that same protein is produced but then immediately sequestered by the cell. The model could help researchers develop a mechanistic description of how living cells achieve precise control of replication and other cellular cycles. The advance could have implications ranging from understanding evolutionary processes to designing synthetic cells.
The phenomenon of multiple, simultaneous rounds of chromosome replication was first understood by Stephen Cooper and Charles Helmstetter in 1968 [2]. The duo showed that the time from replication initiation to cell division is independent of the cell doubling time, meaning that a cell with a doubling time shorter than the replication time must start its replication one or two generations back. But the molecular mechanism that determines when and how replication is initiated remained unknown.
In 1991, Flemming Hansen and colleagues at the Technical University of Denmark proposed a molecular mechanism they termed the initiator titration model [3]. In the model, replication begins when an “initiator” protein, called DnaA, binds to a specific site, called the “origin,” on the chromosome. However, DnaA binds more strongly to other chromosome sites (so-called DnaA boxes). Therefore, replication begins only after the boxes fill up, a process called titration. Titration thus provides a mechanism for controlling the initiation of replication.
We now know that DnaA exists in either an active or an inactive state, and only the active state initiates replication [4]. Because the complexes that activate and deactivate DnaA are produced at various locations on the chromosome, the activation is itself affected by chromosome replication, creating a feedback loop. In principle, this feedback provides a second means of controlling replication initiation. Intriguingly, a recent numerical study by Mareike Berger and Pieter Rein ten Wolde from AMOLF in the Netherlands suggests that neither control mechanism alone, titration or activation, can explain precise initiation for the whole range of physiologically relevant doubling times, but that their combination can [5].
Now Fu and co-workers have upgraded the initiator titration model to include the activation and deactivation of DnaA, as well as other factors identified in recent experiments. By stripping the problem to its essential ingredients, the researchers describe repeated rounds of replication mathematically as a discrete map. An analysis of this map determines the conditions for which replication becomes unstable—meaning a situation with an alternation between two initiations in one cell cycle and none in the next. The result is a phase diagram of replication stability in terms of the key parameters of the problem: the ratio of the chromosome replication time to the cell doubling time, and the ratio of the number of DnaA binding sites at the origin to the number of DnaA boxes. Importantly, the researchers find that including the replication-dependent deactivation of DnaA removes the unstable region of the phase diagram, providing an elegant confirmation of the earlier numerical results [5].
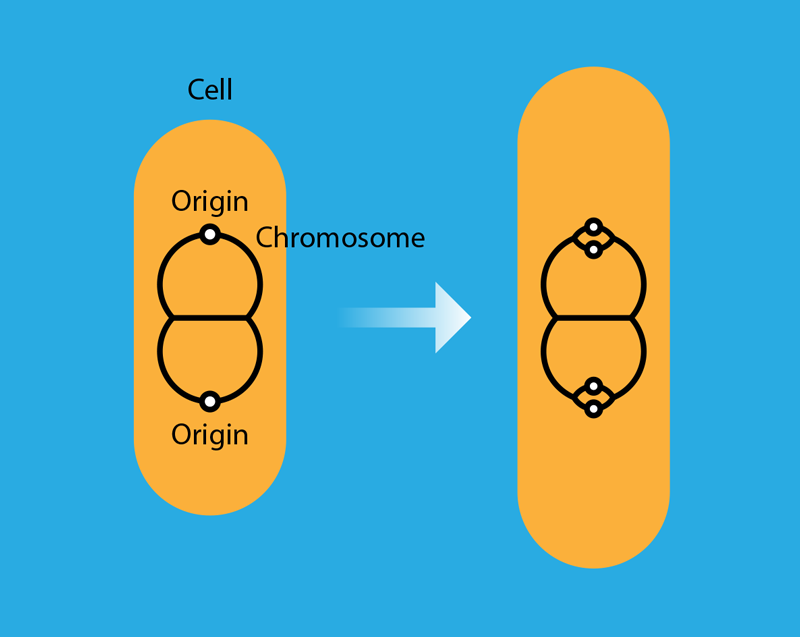
Fu and colleagues also consider the effect of DnaA titration on the degree of synchrony among multiple initiation events in the same cell. When the cell doubling time is faster than the chromosome replication time, two rounds of replication overlap. A replicating chromosome then has two copies of the origin that initiate replication before the cell divides (Fig. 1). These initiation events should be synchronous, that is, occur at about the same time. However, in any given cell, intrinsic noise in the initiation control mechanism leads to some asynchrony. Asynchrony is detrimental: accumulated over multiple generations, it could jeopardize the basic requirement that every cell has a complete copy of the chromosome. The researchers show that without titration, the asynchrony decreases with the number of DnaA binding sites as —a consequence of the Poisson nature of the stochastic process producing the DnaA protein. With titration, initiation becomes a two-step Poisson process: first DnaA binds to the boxes, then it binds to the origins. The first step buffers much of the protein production noise from the second step, and consequently the noise-induced asynchrony decreases more rapidly with , scaling as . In other words, the two-step process boosts the synchrony of multiple initiation events. The synchrony boost offers a simple explanation for why a cell would produce many more copies of DnaA than required to initiate replication, only for them to be sequestered by the DnaA boxes.
The work of Fu and colleagues comes amid a resurgence of interest in bacterial physiology, and many of their predictions may soon be tested. Already, concurrent numerical work by Berger and ten Wolde suggests that titration is indeed necessary for synchronous initiation [6]. Experimentally, decoupling the activation and titration of DnaA is becoming increasingly feasible [7], which may allow researchers to test their predictions on replication stability and to further disentangle the respective roles of the two control mechanisms. More broadly, the control of replication initiation is intimately tied to the control of cell division itself, and the causal relationship between the two is under intense investigation [8].
Precise control of chromosome replication is a fundamental requirement of any growing and dividing cell—and thus fundamental to all of life. What further molecular “innovations” were brought about by evolution to ensure precise replication control? Destabilized control is clearly undesirable, but what are the specific consequences for biological fitness? Could we build synthetic components that replicate as precisely as real ones? Answering these questions is already within sight, and the continued coupling of theory to experiments in this vibrant field will be key to new discoveries.
References
- H. Fu et al., “Bacterial replication initiation as precision control by protein counting,” PRX Life 1, 013011 (2023).
- S. Cooper and C. E. Helmstetter, “Chromosome replication and the division cycle of Escherichia coli Br,” J. Mol. Biol. 31, 519 (1968).
- F.G. Hansen et al., “The initiator titration model: computer simulation of chromosome and minichromosome control,” Res. Microbiol. 142, 161 (1991).
- T. Katayama et al., “The DnaA cycle in Escherichia coli: Activation, function and inactivation of the initiator protein,” Front. Microbiol. 8 (2017).
- M. Berger and P.R. ten Wolde, “Robust replication initiation from coupled homeostatic mechanisms,” Nat. Commun. 13, 6556 (2022).
- M. Berger and P.R. ten Wolde, “Synchronous replication initiation of multiple origins,” PRX Life 1, 013007 (2023).
- A. Knöppel et al., “Regulatory elements coordinating initiation of chromosome replication to the Escherichia coli cell cycle,” Proc. Natl. Acad. Sci. U.S.A. 120 (2023).
- P. Kar et al., “Using conditional independence tests to elucidate causal links in cell cycle regulation in Escherichia coli,” Proc. Natl. Acad. Sci. U.S.A. 120 (2023).