Placing a Full Protein Library Under Pressure
Deep-ocean zones host a vast majority of Earth’s microbial life [1]. These organisms thrive under pressures up to 1000 times that at sea level. Yet the cellular- and molecular-scale adaptations to such extreme conditions remain largely understudied. To shed light on these adaptations, Haley Moran from Johns Hopkins University in Maryland and colleagues introduce high-pressure limited proteolysis (Hi-P LiP), a groundbreaking technique that detects subtle conformational changes in protein structures at pressures that correspond to the deep-sea environment [2]. Unlike previous methods that focus on individual proteins, Hi-P LiP can be used to measure the pressure effects on an organism’s full protein library, or proteome. The researchers apply their technique to a bacterium found in shallow-water hot springs and find that approximately 40% of its proteome undergoes structural changes when exposed to pressures found in the deep sea. The demonstration of the Hi-P LiP technique not only advances our understanding of protein adaptation to high pressures but also holds practical implications for fields such as food science and drug discovery.
Hot springs, acid lakes, and hypersaline lakes host rich and complex ecosystems that have evolved over billions of years to thrive in extreme environments. Similarly, the deep ocean supports microbial and animal life capable of sustaining extremely high hydrostatic pressure, up to 1000 atmospheres (100 MPa) at the deepest ocean points. Understanding how deep biosphere organisms adapt to extreme environments is fundamental to evolutionary biology, as it deepens our knowledge of life’s origins. Additionally, exploring these adaptation mechanisms can drive new discoveries in biotechnology.
Considerable research has focused on how individual biomolecules (proteins, nucleic acids, and lipids) respond to high pressure. These experiments typically involve exposing purified biomolecules to pressure perturbations and monitoring their responses using various biophysical techniques, such as circular dichroism, nuclear magnetic resonance (NMR), fluorescence, infrared spectroscopy, or small angle x-ray scattering [3]. This research has unveiled numerous insights into the structural, dynamical, and functional properties of biomolecules [4]. For lipid assemblies, exposure to high pressure leads to increased rigidity that can cause phase transitions in both single-component lipid bilayers and ternary lipid mixtures [5].
For proteins, researchers have studied how these complex molecules change shape under extreme pressures. Between 100 and 200 MPa, proteins tend to adopt compact conformations that resemble smaller-sized versions of their native biologically active state. These alternate conformations often represent low-lying excited states of the protein that may have important functional roles. At even higher pressures (greater than 300 MPa), most proteins will partially or fully unfold. This pressure-induced unfolding is driven by the penetration of water molecules into the protein core, disrupting hydrophobic contacts [6]. Once those contacts are broken, the protein opens up—a response that is explained by the fact that the molecule’s unfolded state generally occupies a smaller molar volume than its folded state. Several biophysical studies have shown that the volume difference between these states primarily arises from cavities and small voids, which are part of the folded structure but become fully hydrated as the protein unfolds [7–9].
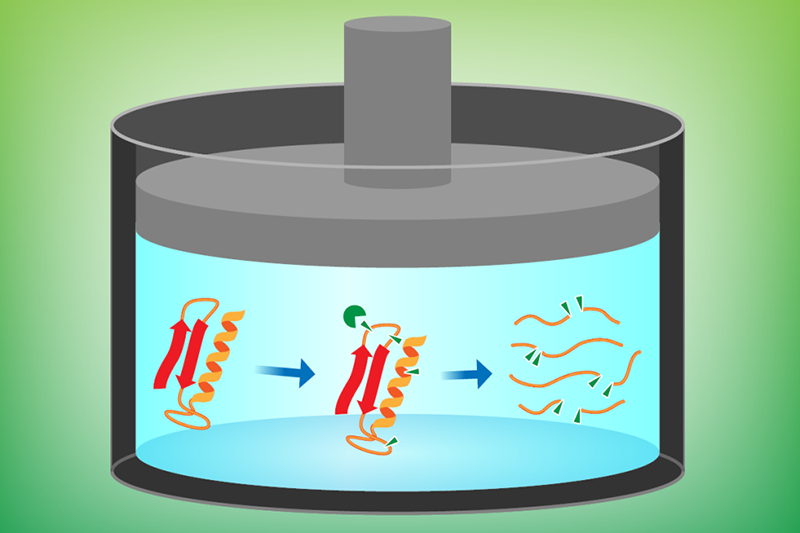
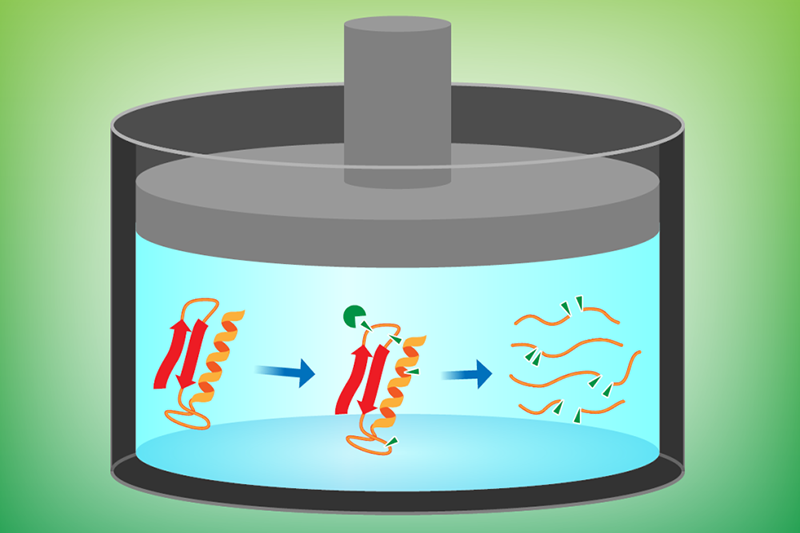
Previous studies have typically worked with isolated proteins in controlled in vitro environments—an approach that is susceptible to selection bias and is time-consuming in terms of preparation. Moran and colleagues developed their Hi-P LiP method to detect structural changes occurring within a complete set of bacterial proteins that can be obtained directly from cell extracts. The technique involves using a proteolytic enzyme (Proteinase K) to cleave proteins at specific sites, followed by mass spectrometry to identify and analyze the resulting fragments (Fig. 1).
As a first demonstration, the researchers investigated how pressure affects the proteins of Thermus thermophilus, a form of bacteria that lives in hot springs. The organism’s entire proteome was analyzed first at its natural habitat pressure (0.1 MPa) and then at a deep-ocean-mimicking pressure (100 MPa). The research team found that nearly 40% of T. thermophilus proteins exhibit significantly different patterns of cleavage sites at low and high pressures. The reason for this difference is that sites hidden at atmospheric pressure from the cleaving enzyme become exposed—and thus cut—as a result of conformational changes experienced by these proteins at high pressure.
This innovative method allows for detailed examination of pressure-induced structural changes, providing unprecedented insight into the adaptability of proteins under hydrostatic pressure. One of the most intriguing findings of this study is that proteins with lower packing density—those that contain more empty space within their structure—are unexpectedly more resistant to deformation when subjected to high pressure. These findings seem to challenge biophysics-based models, which assume that these less compact proteins should be more likely to unfold under pressure [5]. However, Moran and colleagues argue that there may not be any contradiction: The Hi-P LiP method is sensitive to local deformations that occur before complete unfolding. Thus, the less compact proteins may be more resistant to deformations (more rigid), but once the pressure gets high enough, they are the first to unfold.
As for other interesting results, the researchers found that proteins that are rich in either acidic or basic amino acids have a higher propensity for structural alterations under pressure than proteins containing an equal mix of acidic and basic amino acids. They also observed that proteins with disordered regions are more prone to pressure-induced alterations, although the conformational changes detected by Hi-P LiP are not necessarily located within disordered regions themselves.
This work opens up exciting possibilities for future research in structural biology and biophysics. Now that the effectiveness of Hi-P LiP in studying protein behavior under high pressure has been demonstrated, researchers can apply this method to other extremophiles and various proteomes, further unraveling the mysteries of life in extreme environments. This could lead to new discoveries about protein stability, function, and adaptability, offering valuable insights that can be harnessed in biotechnology applications, such as designing more resilient enzymes for industrial applications or improving the effectiveness of therapeutic proteins.
References
- C. Corinaldesi, “New perspectives in benthic deep-sea microbial ecology,” Front. Mar. Sci. 2 (2015).
- H. M. Moran et al., “Proteome-wide assessment of protein structural perturbations under high pressure,” PRX Life 2, 033011 (2024).
- L. Smeller, “Biomolecules under pressure: Phase diagrams, volume changes, and high pressure spectroscopic techniques,” Int. J. Mol. Sci. 23, 5761 (2022).
- J. L. Silva et al., “High-pressure chemical biology and biotechnology,” Chem. Rev. 114, 7239 (2014).
- R. Winter, “Effects of hydrostatic pressure on lipid and surfactant phases,” Curr. Opin. Colloid Interface Sci. 6, 303 (2001).
- J. Roche and C. A. Royer, “Lessons from pressure denaturation of proteins,” J. R. Soc., Interface. 15, 20180244 (2018).
- K. J. Frye and C. A. Royer, “Probing the contribution of internal cavities to the volume change of protein unfolding under pressure,” Protein Sci. 7, 2217 (1998).
- J. Roche et al., “Cavities determine the pressure unfolding of proteins,” Proc. Natl. Acad. Sci. U.S.A. 109, 6945 (2012).
- N. V. Nucci et al., “Role of cavities and hydration in the pressure unfolding of T4 lysozyme,” Proc. Natl. Acad. Sci. U.S.A. 111, 13846 (2014).