The Aquatic Dance of Bacteria
Bacteria are among the oldest and most abundant living species on Earth, and their activity influences the planet’s environmental dynamics in multiple ways. Microbial metabolic products maintain soil structure, marine bacteria control the biochemistry and photosynthetic productivity of the oceans [1], and Geobacter bacteria are able to clean contaminated sites by “feeding” on heavy metals such as uranium [2]. Bacteria often migrate en masse over large distances, moving in dense groups in a highly organized, collective fashion known as “swarming motility.” Compared to independent individual movements, such collective motions have multiple evolutionary advantages: a bacterial colony can more easily populate new territories, search for food, and increase its survival probability in harsh conditions. Swarming is also an effective strategy to prevail against antibiotics [3].
The flow dynamics of dense bacterial colonies can be very complex and, because of the interaction between the bacteria and the fluid, remarkably different from those predicted by conventional fluid models. In particular, turbulent swimming patterns often emerge, characterized by chaotic motions and the formation of vortices, even in situations where liquids should exhibit laminar flow. But these complex phenomena are difficult to characterize experimentally, and a predictive model that describes them has not emerged to date. Now, as reported in Physical Review Letters, Jörn Dunkel at the University of Cambridge, UK, and co-workers [4] have carried out experiments capable of separately measuring the movements of the fluid and those of the suspended bacteria, and have developed a minimal theory that quantitatively captures their experimental observations.
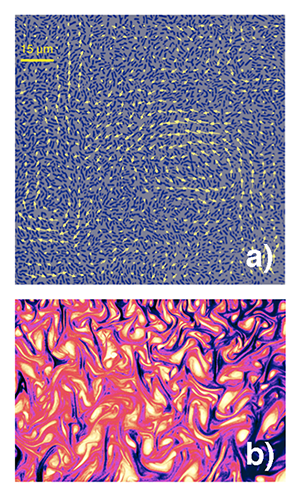
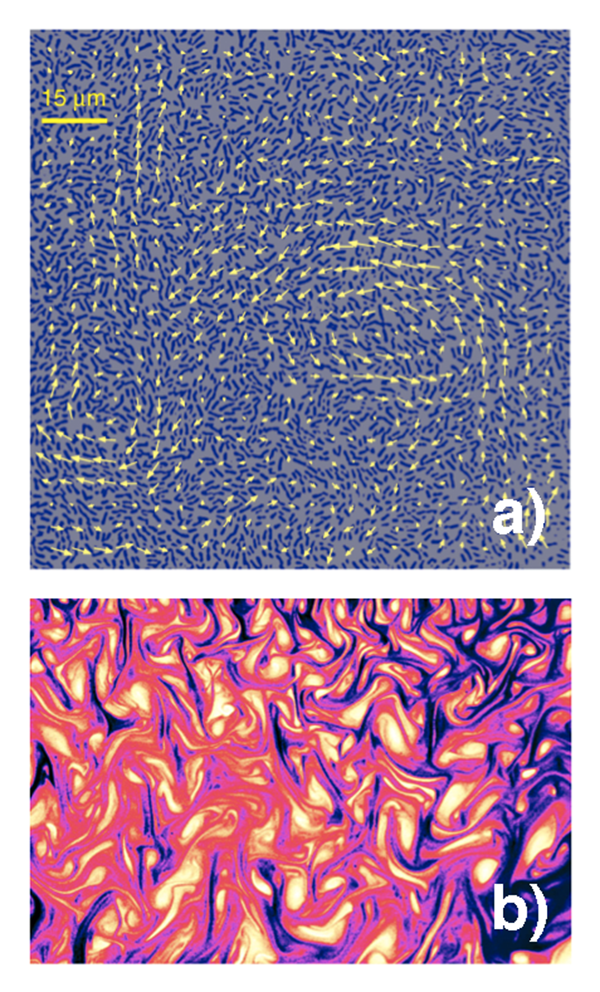
Assemblies of self-propelled organisms, from large animal swarms to microscopic bacterial colonies, are often termed “active fluids” and the study of their collective dynamics has attracted enormous attention in the last decade [5]. Fluids containing swimming bacteria are ideal model systems for such studies: bacteria are relatively simple compared to other living organisms, can be grown in large quantities, and experiments can be performed under controlled conditions. Concentrated solutions of bacteria, such as the common bacteria Bacillus subtilis (also known as hay or grass bacillus), show remarkable patterns of collective motion [6], manifested by the formation of self-organized jets and vortices significantly exceeding the size of a single bacterium [Fig. 1(a)]. While this motion is visibly similar to familiar turbulence in vigorously stirred liquids [see Fig. 1(b)], there are important differences. In hydrodynamic turbulence the mechanical energy is injected at the macroscopic scale, e.g., by stirring the liquid, which then trickles down to the microscale. In this case, turbulence can only kick in when inertial forces dominate viscosity, e.g., when the Reynolds number (the ratio of inertial to viscous forces) is sufficiently high (generally ). In contrast, bacteria live in an extremely viscous environment: typical Reynolds numbers are on the order of – .
At such numbers in a conventional liquid, one would expect laminar flow. But in bacterial turbulence, the energy is injected at the microscopic scale by the beating of bacterial flagella, which makes the physical properties of this type of turbulence fundamentally different from the turbulent flow of liquid. The current consensus is that this form of collective motion is determined by a few pure physical mechanisms (contrasted to biological chemotaxis—the motion due to gradients of certain chemicals), such as the hydrodynamic coupling of bacteria induced by self-generated flow and direct collisions. Although a number of theoretical concepts have been proposed to address various aspects of collective motion [7–10], the importance of these mechanisms is still under discussion [11].
In bacterial turbulence, the movement of bacteria is strongly coupled to the flow of the liquid: a swimming bacterium influences the neighbors by creating flow in the viscous liquid; in turn, the fluid draws the bacteria along. This makes it difficult to disentangle the role of bacterial motion from that of the fluid flow in experiments. Now, Dunkel et al. have been able to probe the fluid and the bacterial dynamics independently by performing two complementary measurements in succession on the same bacterial colony. First, they applied bright-field microscopy to probe the movement of the bacteria. Second, they added micron-sized fluorescent markers to the suspension, small enough so as not to distort the flow, and traced the trajectories of the markers by fluorescence microscopy to visualize the flow of the suspending fluid.
This novel combination of experimental techniques yielded a number of interesting observations. The authors were able to probe the dynamics of the active fluid over a two-order-of-magnitude range of kinetic energy, by exploiting the fact that the activity of bacteria (i.e., their swimming speed) depends on how much oxygen they can breathe; the energy released by bacteria into the liquid thus decreased as the experiment progressed and oxygen was consumed. Remarkably, over this whole range the relation between the kinetic energy and dissipation (quantified by the enstrophy, or mean-squared flow vorticity) was linear. Further, the spatial correlations in the velocity of bacteria and liquid showed only a weak dependence on the activity of bacteria, thus reinforcing the earlier finding of a study in which I participated [11] that the properties of collective motion (e.g., the typical vortex size) do not depend on the amount of energy injected by bacteria into the liquid. Finally, the experiments suggested that for high concentrations of bacteria, the statistics of the flow and the motion of bacteria are tightly linked (although it cannot be fully excluded that some of the fluorescent markers are simply “pushed” by the bacteria rather than drawn along by the fluid).
The authors interpret their experimental results within the framework of a minimal phenomenological model they previously introduced [10]. The model incorporates the most relevant physical effects: incompressibility of the fluid, formation of jets by local alignment of bacteria via collisions, and the propensity to generate vortices of a certain size (so-called short-wave instability). Simulations based on the model, with appropriate parameter choice, closely reproduce many experimental findings, like the shape of the correlation functions, the flow structure, etc. The model also allows reconstructing quantities that are not directly visible in the experiments, such as the three-dimensional distribution of the vorticity.
Although the phenomenological model fits the data well, one can point to a theoretical shortcoming that will motivate further work. The model is inspired by an equation for the so-called short-wave instability proposed by Swift and Hohenberg in 1977 [12] to describe hydrodynamic fluctuations in Rayleigh-Bénard convection (liquids heated from below). The equation describes how, in a uniform state, a short-wave instability can arise when spatial modulations with an optimal characteristic scale grow exponentially (in Rayleigh-Bénard convection, such optimal scale is determined by height of the liquid layer). The simplest (i.e., cubic) nonlinear term is also introduced to limit the growth.
Here comes the catch: one can show that, except in a few very special cases [13], an equation of this type cannot be asymptotically derived (i.e., derived as the mathematical limit for certain parameters value) from the set of underlying equations that microscopically describe the active fluid. Namely, the expansion can be carried out with respect to the amplitude of small spatial modulation, but not with respect to small spatial gradients—the short-wave instability sets a finite spatial scale. In essence, the equation with only the simplest nonlinear terms cannot be generally obtained; inevitably, further models will need to be developed. Can one develop similar phenomenological models that can be rigorously connected to the microscopic description of self-propelled particles interacting with suspending fluid?
The work of Dunkel et al. clearly shows the possibility of probing bacterial and fluid motion independently and demonstrates that a simple phenomenological model may successfully describe complex dynamic phenomena such as bacterial turbulence. This will certainly provide food for thought to experimenters and theorists wishing to understand how the emergence of collective behavior affects the dynamics of bacterial motions and of active fluids in general.
References
- R. Stocker, “Marine Microbes See a Sea of Gradients,” Science 2, 628 (2012)
- D. L. Cologgi, S. Lampa-Pastirk, A. M. Speers, Sh. D. Kelly, and G. Reguera, “Extracellular Reduction of Uranium via Geobacter Conductive Pili as a Protective Cellular Mechanism,” Proc. Natl. Acad. Sci. U.S.A. 108, 15248 (2011)
- M. T Butler, Q. Wang, and R. M. Harshey, “Cell Density and Mobility Protect Swarming Bacteria Against Antibiotics,” Proc. Natl. Acad. Sci. U.S.A. 107, 3776 (2009)
- J. Dunkel, S. Heidenreich, K. Drescher, H. H. Wensink, M. Bär, and R. E. Goldstein, “Fluid Dynamics of Bacterial Turbulence,” Phys. Rev. Lett. 110, 228102 (2013)
- T. Vicsek and A. Zafeiris, “Collective Motion,” Phys. Rep. 517, 71 (2012)
- C. Dombrowski, L. Cisneros, S. Chatkaew, R. E. Goldstein, and J. O. Kessler, “Self-Concentration and Large-Scale Coherence in Bacterial Dynamics,” Phys. Rev. Lett. 93, 098103 (2004)
- D. Saintillan and M. Shelley, “Instabilities and Pattern Formation in Active Particle Suspensions: Kinetic Theory and Continuum Simulations,” Phys. Rev. Lett. 100, 178103 (2008)
- D. L. Koch and G. Subramanian, “Collective Hydrodynamics of Swimming Microorganisms: Living fluids,” Annu. Rev. Fluid. Mech. 43, 637 (2011)
- I. S. Aranson, A. Sokolov, J. O. Kessler, and R. E. Goldstein, “Model for Dynamical Coherence in Thin Films of Self-Propelled Microorganisms,” Phys. Rev. E 75, 040901 (2007)
- H. H. Wensink, J. Dunkel, S. Heidenreich, K. Drescher, R. E. Goldstein, H. Löwen, and J. M. Yeomans, “Meso-scale Turbulence in Living Fluids,” Proc. Natl. Acad. Sci. U.S.A. 109, 14308 (2012)
- A. Sokolov and I. S. Aranson, “Physical Properties of Collective Motion in Suspensions of Bacteria,” Phys. Rev. Lett. 109, 248109 (2012)
- J. Swift and P. C. Hohenberg, “Hydrodynamic Fluctuations at the Convective Instability,” Phys. Rev. A 15, 319 (1977); M. C. Cross and P. C. Hohenberg, “Pattern Formation out of Equilibrium,” Rev. Mod. Phys. 65, 851 (1993)
- I. S. Aranson and L. Kramer, “The World of the Complex Ginzburg-Landau Equation,” Rev. Mod. Phys. 74, 99 (2002)