How Viruses Get Their Protective Shells
Viruses encase their genetic material in a protective protein shell known as a capsid. Despite rapid advances in both theoretical and experimental biology, the processes by which capsids form have remained uncertain. This knowledge gap has impeded the development of antiviral treatments that inhibit capsid assembly and of artificial capsids for use in targeted drug delivery. Now a computational analysis by Banu Ozkan and her colleagues at Arizona State University has revealed what could be a general mechanism for capsid formation [1].
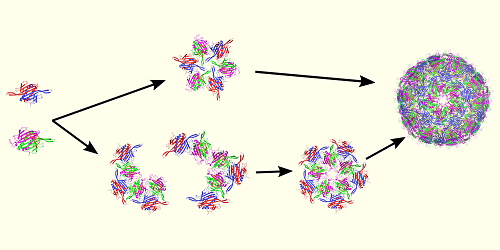
Ozkan and her colleagues considered the virus MS2, which has a simple structure and has been extensively studied. MS2 infects the intestinal bacterium Escherichia coli, and its genetic material is encoded in single strands of the molecule RNA. The researchers explored capsid formation in MS2 using two computational methods. The first tracked the flexibility of the developing capsid’s amino acids—the building blocks that make the proteins in this structure. The second tracked how strongly each of those amino acids coupled to the others.
Ozkan and her colleagues found that, in their simulations, the capsid assembly in MS2 followed a sequential pathway that was driven by changes in the dynamics of the forming capsid’s amino acids. The pathway involved a series of steps in which amino acids assembled into small structures that then assembled into larger structures to make the capsid. These dynamics were regulated by interactions between those amino acids and the virus’s RNA. The researchers say that, although their study focused on a simple virus, its implications might extend to more complex viruses—such as SARS-CoV-2, which causes COVID-19. They also say that similar mechanisms could be at work in other protein–RNA complexes, including ribosomes, the protein manufacturing sites in cells.
–Ryan Wilkinson
Ryan Wilkinson is a Corresponding Editor for Physics Magazine based in Durham, UK.
References
- S. Hamilton et al., “RNA-induced allosteric coupling drives viral capsid assembly,” PRX Life 2, 013012 (2024).