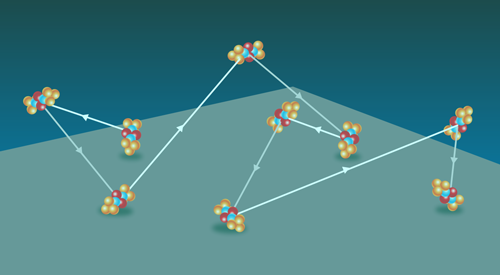
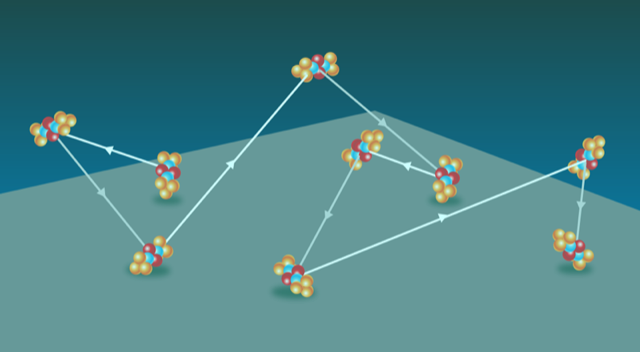
3D Imaging of Hopping Molecules
Theory predicts that the movement of molecules along the surface of a cell is dominated by “hops,” with the molecules jumping in and out of the liquid surrounding the cell [1, 2]. Such a process, for example, is thought to occur when molecules transport cargoes from one region of the cell to another. But experimentally tracking the full 3D motion of such molecules is notoriously difficult, making it hard to confirm these predictions. Now, Daniel Schwartz from the University of Colorado Boulder and colleagues have done exactly that, capturing the 3D trajectories of human serum albumin molecules on silica surfaces submersed in a water-glycerol mixture [3]. The team observed that the strength of the electrostatic interactions between the surface and the molecules strongly influence the characteristics of the hops, such as their length and duration. The research shows the potential of fluorescence imaging for tracking and probing the 3D motion of single molecules along a submersed surface. Such an ability could enable a deeper understanding of molecular processes in cells.
The path of a molecule through a fluid typically resembles a random walk due to its random (Brownian) motion, a behavior that is well understood. But add in an interface, like the surface of a cell, and things get more complicated. Theory predicts that molecules wouldn’t simply track a random path along the cell’s surface. Rather, their paths are interrupted by hops off the surface and into the liquid surrounding the cell. Experimental results lend support to this prediction, but to date, they have not directly determined the molecules’ paths in 3D. As such, the details of the molecules’ motion and the influence of the surface on this motion remain unknown. Do the molecules move predominantly along the surface or in the liquid? Or is their time divided equally between both? How big are the hops and how long do they last? How does the molecules’ motion depend on the strength of their interactions with the fluid and the surface? Knowing the answers to these questions would improve researchers’ understanding of the diffusion of molecules in such situations, and it would give insights into the physical, chemical, and biological processes to which the molecules contribute [4–6].
Schwartz’s team takes a step towards answering these questions using an adaption of a recently developed fluorescence imaging technique [7]. In standard fluorescence imaging, molecules are tagged with a dye, which fluoresces when illuminated with light of a specific wavelength. The fluorescence serves as a tiny torch reporting the whereabouts of the particle. In cases where the molecule is moving, the fluorescence can be used to track the particle and map its trajectory in space and time.
Typically, fluorescence images consist of 2D projections of 3D positions, in the same way that photographs taken with our cell phones are 2D projections of a 3D scene. However, unlike photographs, fluorescence images contain only a series of light spots, which provide little perspective information for the viewer. Additionally, noise in images and the fact that the light spots are generally much bigger than the molecules themselves limit the ability of researchers to accurately determine the location of a molecule in the vertical direction—its height above the surface—using this technique.
The technique of Schwartz and colleagues [3] circumvents this obstacle by determining a molecule’s changing vertical position using an optical method to modify its so-called double-helix point spread function. Adding a phase-filter to the optical path of a fluorescence microscope, the team modified the light from the fluorescence emission such that two spots, not one, appear on the microscope’s camera. As the molecule’s height changes, the two spots rotate around each other in the manner of a double helix, hence the name of the point spread function. The exact angle of rotation depends on the molecule’s height. In this way, the team mapped the 3D motion of a molecule with a spatial precision of 20 nm in the vertical direction and 15 nm in the two lateral directions, and with a temporal precision of around 0.1 s.
Tagging individual human serum albumin molecules with a fluorescent dye, Schwartz and colleagues captured the trajectories of individual molecules moving along negatively charged silica surfaces (Fig. 1) that were submersed in a water-glycerol liquid. The researchers modified the surfaces with various silane compounds to tune the electrostatic interactions between the molecules and the silica surface. The team observed hops and, in some cases, “flights” of the molecules as they moved across the surface.
Analyzing the statistics of the observed hops, the team found that the “stickiness” of the surface—how many jumps a molecule makes before being absorbed back to the surface—depends on the strength of the molecule-surface electrostatic interaction. Molecules moving on the most attractive type of surface (amino-silane coated silica) hopped twice on average, while molecules moving on the most repulsive type of surface (uncoated fused silica) kept going for seven jumps. The team found that the distribution of the waiting times between jumps increased with the attractive interaction strength of the surface, a behavior not predicted for simple Brownian motion [8]. The nature of the interaction (repulsive or attractive) also influenced the length, height, and duration of the hops—molecules moving on more repulsive surfaces had longer, higher, slower jumps due to the pushing away of the molecule by the surface. As such, the motion of the molecules no longer fits that of simple Brownian motion but is instead biased by the surface, potentially allowing the molecules to move faster or slower.
Faster diffusion could be beneficial for myriad cellular processes involving the transport of molecules over the cell’s outer membrane and over the membrane surrounding the cell nucleus (nuclear envelope). Imagine pedestrians walking in an overcrowded city. If some had an added ability to jump into the third dimension, they could leapfrog those walking more slowly. Such an action could dramatically enhance their mobility. Similarly, a hopping molecule searching for a target on the cell’s surface could carry out a more efficient search than a molecule that only moves in 2D, as it could continually hop to new, unsearched parts of the cell without having to resample any areas. Thus the experiments show the important contribution of interactions between molecules and surfaces in determining the dynamics of the molecules, something that is not yet fully understood. Learning more about the dynamics of molecules could provide a more detailed picture of the behavior of proteins and other biomolecules in processes including the trafficking of nutrients into the cell, the signaling of information to the cell nucleus, and the initiation of cell division.
This research is published in Physical Review Letters.
References
- O. V. Bychuk, B. O'Shaughnessy, “Anomalous Diffusion at Liquid Surfaces,” Phys. Rev. Lett. 74, 1795 (1995).
- A. V. Chechkin, I. M. Zaid, M. A. Lomholt, I. M. Sokolov, and R. Metzler, “Bulk-Mediated Diffusion on a Planar Surface: Full Solution,” Phys. Rev. E 86, 041101 (2012).
- D. Wang, H. Wu, and D. Schwartz, “Three-Dimensional Tracking of Interfacial Hopping Diffusion,” Phys. Rev. Lett. 119, 268001 (2017).
- E. Barkai, Y. Garini, and R. Metzler, “Strange Kinetics of Single Molecules in Living Cells,” Phys. Today 65, No. 8, 29 (2012).
- C. Manzo and M. F Garcia-Parajo, “A Review of Progress in Single Particle Tracking: from Methods to Biophysical Insights,” Rep. Prog. Phys. 78, 124601 (2015).
- D. Krapf, G. Campagnola, K. Nepal, and O. B. Peersen, “Strange Kinetics of Bulk-Mediated Diffusion on Lipid Bilayers,” Phys. Chem. Chem. Phys. 18, 12633 (2016); G. Campagnola, K. Nepal, B. W. Schroder, O. B. Peersen, and D. Krapf, “Superdiffusive Motion of Membrane-Targeting C2 Domains,” Sci. Rep. 5, 17721 (2015).
- S. R. P. Pavani, M. A. Thompson, J. S. Biteen, S. J. Lord, N. Liu, R. J. Twieg, R. Piestun, and W. E. Moerner, “Three-Dimensional, Single-Molecule Fluorescence Imaging Beyond the Diffraction Limit by Using a Double-Helix Point Spread Function,” Proc. Natl. Acad. Sci. U.S.A. 106, 2995 (2009); See also W. E. Moerner, Nobel Prize lecture, https://www.nobelprize.org/mediaplayer/index.php?id=2411.
- E. Schrödinger, “Zur Theorie der Fall- und Steigversuche an Teilchen mit Brownscher Bewegung,” Z. Phys. 16, 289 (1915); See also S. Redner, A Guide to First-Passage Processes (Cambridge University Press, Cambridge, 2007)[Amazon][WorldCat].